Drug Designing: Open Access
Open Access
ISSN: 2169-0138
ISSN: 2169-0138
Research Article - (2021)Volume 10, Issue 1
The Covid-19 pandemic with 74,299,042 confirmed cases including 1,669,982 deaths, reported worldwide by WHO (accessed 1:21 PM EST on 12/20/2020) is one of the most challenging situations the humankind is facing currently. Though some vaccines to treat this catastrophic disease have been developed on fast track, but still exploring the more potential alternative therapies is need of the hour. In search of new drug candidates to treat this pandemic, a structure based virtual screening study was carried out on the 125 compounds isolated from the native Caribbean medicinal plants like Aloe vera, Lemon grass, Moringa Olifera, and Lignum vitae, to determine their inhibitory potential against three main drug targets of SARS CoV-2; main protease (PDB ID: 6LZE), nsp 15 (PDB ID: 6WLC), and RNA-dependent RNA Polymerase (PDB ID: 7BV2) of SARS-CoV-2. The virtual screening workflow was performed using Schrodinger small molecule drug discover suite, in that compounds were prepared and docked in active sites of all three proteins. The co-crystallized ligand of every protein was used as a positive control and all docking scores were compared with it. Out of 125 compounds, 37 showed favorable docking interaction with the proteins compared to the native ligand. 13 compounds showed excellent docking score against 6LZE, out of that the most negative docking score (-11.962) was of rutin in compare to native ligand (-8.095), and it also showed more negative Prime MMGBSA binding energy-70.54 kcal/mol. In case of 7BV2, 35 compounds showed better docking results than the native ramdesivir (-4.566), docking score (-8.194) of vicenin-2 was best, it also exhibited almost same binding energy of complex (-44.54 Kcal/mol) as ramdesivir. In case of 6 WLC, in site 1, orientin showed most negative dockings core (-10.896), while in site 2 swertiajaponin’s docking score is-12.364. Pharmacokinetic (ADME) properties of 37 compounds were also calculated to ascertain the druglikeness of potential inhibitors. This study provides some lead molecules that might be helpful in designing drug for treating corona virus disease.
Main protease; NSP-15: Nonstructural protein-15; RNA dependant RNA polymerase; Caribbean medicinal plants; Aloe vera; Lemon grass; Moringa Olifera; Lignum vitae
The novel coronavirus SARS CoV-2, better known as COVID-19, is a new strain that belongs to the SARS family. This family of viruses replicates by binding to ACE receptors of cells and entering them to replicate. The virus itself consists of 2 subunits, one which binds to the ACE receptor and the other than binds to the cell surface to anchor the virus. The virus is most commonly found in a mass population among animals however, the new strain, SARS CoV-2 has been able to thrive among humans. A study of the genomic structure of the different coronavirus strains found that the new strain shares 88% similarity with the bat strain of SARS [1]. Reports state that the main mode of transmission for the SARS CoV-2 virus is through person-to-person contact specifically by way of droplets from an infected person coming into contact with a healthy host.
The trend of infection and symptom severity is directly linked to immune health whereby immunocompromised persons have a higher risk of infection and severe symptoms. Currently, 14 days is the known incubation period of the virus however there have been cases where the virus showed symptoms earlier or later than expected [2]. The main target of the virus is the respiratory system. As is common among the coronavirus family, symptoms presented include a dry coughing, fever, and fatigue however this new strain presented symptoms particular to the virus [3]. In addition to the aforementioned, SARS CoV-2 also caused disruption as the lower airway such as sneezing and sore throat in addition to gastrointestinal symptoms which are rare for the coronavirus family [3].
Due to how infectious the virus is, social distancing and mask regulations have been implemented leading to the shutdown or heavy patron restriction of closed spaces. Because of this disruption in day to day life, treatment for or immunity to the virus is necessary. Common methods of drug development take an extensive amount of time which is what makes computer-aided drug design a favorable computational tool. Computer-aided drug design is a computation drug design method that involves identifying active sites of target proteins and simulating protein- compound interactions to find inhibitors. The two types of computational drug design are structure-based drug design and ligand-based drug design [4]. Ligand-based drug design is used when the ligand structure is known but the receptor structure is unknown. Structure-based drug design on the other hand involves analyzing the known receptor structure to identify potential active sites, if unknown, and simulate ligand interactions at those sites to identify potential inhibitors that can compete with the biological ligand [4].
There are different proteins that are associated with SARS CoV- 2 functionality that are potential drug targets. Inhibition of these proteins could render the virus ineffective as interactions and mechanisms would be disrupted. The main protease (Mpro) is an essential drug target which, along with papain-like proteases catalyzes the processing of polyproteins translated from viral RNA and recognizes specific cleavage sites [5]. The RNA dependent RNA polymerase (RdRp) has been evident to possess nsp (7,8,12); each RdRp acts as responsive cofactors when stimulating polymerase complex activities. The CoV nsp7/nsp8/nsp12 complex constitutes nucleotide polymerization for its configuration [6]. In addition, the nsp 15 (active hexamer) is responsible for providing structural attributes for the development of innovative therapeutic agents [7]. The SARS-CoV-2 polyprotein assists with transportation mechanisms functioning with maintenance, transcription, and replication of the virus genome.
In this study, four medicinal plants aloe vera, lemon grass, Moringa oleifera and Lignum vitae native to the Caribbean were chosen to determine the inhibitory effects of the antiviral compounds isolated from them on different SARS CoV-2 proteins. Based on the literature [8-11] a list of 125 compounds were compiled in Table 1 and were virtually docked in the active sites of Main protease, RNA dependent RNA Polymerase, and nsp-15 proteins of SARS- CoV-2 by using molecular docking program (GLIDE, Schrodinger). Docking scores and binding energies of protein-ligand complexes were compared with the native ligand of the chosen protein structures. ADME properties of selected compounds were also determined.
| S. No. | Compounds | Plant Origin |
|---|---|---|
| 1 | Chrysophanol | Aloe Vera |
| 2 | C-2'-decoumaroyl-aloeresin G | Aloe Vera |
| 3 | Aloe emodin | Aloe Vera |
| 4 | Aloe C-glucosylchromone | Aloe Vera |
| 5 | Aloin | Aloe Vera |
| 6 | 4-methylpent-3-en-1-ol | Aloe Vera |
| 7 | 4,7-dihydroxy-5-methylcoumarin | Aloe Vera |
| 8 | Aloe emodin anthrone | Aloe Vera |
| 9 | Ruxolitinib phosphate | Aloe Vera |
| 10 | Ruxolitinib | Aloe Vera |
| 11 | Aloesin | Aloe Vera |
| 12 | 6-Methyl-1,3,8-trihydroxyanthraquinone (emodin) | Aloe Vera |
| 13 | Luteolin-8-C-glucoside (orientin) | Aloe Vera |
| 14 | Feruloylquinic acid | Aloe Vera |
| 15 | 10-Hydroxyaloin A | Aloe Vera |
| 16 | Isovitexin | Aloe Vera |
| 17 | Isoaloeresin D | Aloe Vera |
| 18 | Aloin B | Aloe Vera |
| 19 | 5,3′-Dihydroxy-6,7,4′-trimethoxyflavone (eupatorin) | Aloe Vera |
| 20 | Trihydroxy octadecenoic acid | Aloe Vera |
| 21 | 3,4-Di-O-caffeoylquinic acid | Aloe Vera |
| 22 | 6-methylhept-5-en-2-one | Lemon Grass |
| 23 | Camphene | Lemon Grass |
| 24 | Limonene | Lemon Grass |
| 25 | Nonan-4-ol | Lemon Grass |
| 26 | Citronellal | Lemon Grass |
| 27 | Citronellol | Lemon Grass |
| 28 | Neral | Lemon Grass |
| 29 | Geraniol | Lemon Grass |
| 30 | Citral | Lemon Grass |
| 31 | Geranyl acetate | Lemon Grass |
| 32 | β-caryophyllene | Lemon Grass |
| 33 | γ-muurolene | Lemon Grass |
| 34 | Caryophyllene oxide | Lemon Grass |
| 35 | Swertiajaponin | Lemon Grass |
| 36 | 7-epi-ent-eudesmane-5,11-diol | Lemon Grass |
| 37 | (2E,6E)-hedycaryol | Lemon Grass |
| 38 | 1,2-Benzenedicarboxylic acid,mono(2-ethylhexyl) ester | Moringa Oleifera |
| 39 | 1,3-dibenzyl urea | Moringa Oleifera |
| 40 | 2-Ethyl-2-propyl-1-hexanol | Moringa Oleifera |
| 41 | 3,3-dimethyloctane | Moringa Oleifera |
| 42 | 4- Dodecanol | Moringa Oleifera |
| 43 | 4-(-L-rhamnopyranosyloxy) benzyl glucosinolate (glucomoringin) | Moringa Oleifera |
| 44 | 4,6,8-Trimethyl-1-nonene | Moringa Oleifera |
| 45 | All-E-lutein | Moringa Oleifera |
| 46 | All-E-Zeaxanthin | Moringa Oleifera |
| 47 | -Phellandrene | Moringa Oleifera |
| 48 | Apigenin | Moringa Oleifera |
| 49 | Arachidic acid | Moringa Oleifera |
| 50 | Astragalin | Moringa Oleifera |
| 51 | Aurantiamide acetate | Moringa Oleifera |
| 52 | Behenic acid | Moringa Oleifera |
| 53 | Benzyl glucosinolate (glucotropaeolin) | Moringa Oleifera |
| 54 | Benzylamine | Moringa Oleifera |
| 55 | β-sitosterol | Moringa Oleifera |
| 56 | Caffeic acid | Moringa Oleifera |
| 57 | Chlorogenic acid | Moringa Oleifera |
| 58 | Cryptochlorogenic acid | Moringa Oleifera |
| 59 | D-allose | Moringa Oleifera |
| 60 | Dibutyl phthalate | Moringa Oleifera |
| 61 | Ellagic acid | Moringa Oleifera |
| 62 | Epicatechin | Moringa Oleifera |
| 63 | Eugenol | Moringa Oleifera |
| 64 | Ferulic acid | Moringa Oleifera |
| 65 | Gallic acid | Moringa Oleifera |
| 66 | Genistein | Moringa Oleifera |
| 67 | Gentisic acid | Moringa Oleifera |
| 68 | Glucoconringiin | Moringa Oleifera |
| 69 | Glucoconringiin(1−) | Moringa Oleifera |
| 70 | Hentriacontane | Moringa Oleifera |
| 71 | Hexanedioic acid, bis (2-ethylhexyl) | Moringa Oleifera |
| 72 | Isoquercetin | Moringa Oleifera |
| 73 | Isorhamnetin | Moringa Oleifera |
| 74 | Kaempferol | Moringa Oleifera |
| 75 | Kaempferol-3-O--rhamnoside | Moringa Oleifera |
| 76 | Kaempferol-3-O-glucoside | Moringa Oleifera |
| 77 | Kaempferol-3-rutinoside | Moringa Oleifera |
| 78 | Linoleic acid | Moringa Oleifera |
| 79 | Linolenic acid | Moringa Oleifera |
| 80 | Luteolin | Moringa Oleifera |
| 81 | Marumoside A | Moringa Oleifera |
| 82 | Marumoside B | Moringa Oleifera |
| 83 | Moringyne | Moringa Oleifera |
| 84 | Multiflorin B | Moringa Oleifera |
| 85 | Myricetin | Moringa Oleifera |
| 86 | Myristic acid | Moringa Oleifera |
| 87 | N--L-rhamnopyranosyl vincosamide | Moringa Oleifera |
| 88 | Niazimicin | Moringa Oleifera |
| 89 | Niaziminin | Moringa Oleifera |
| 90 | Niazirin | Moringa Oleifera |
| 91 | Niazirinin | Moringa Oleifera |
| 92 | o-Coumaric acid | Moringa Oleifera |
| 93 | O-ethyl-4-[(-L-rhamnosyloxy)-benzyl] carbamate | Moringa Oleifera |
| 94 | Oleic acid | Moringa Oleifera |
| 95 | p-Coumaric acid | Moringa Oleifera |
| 96 | p-Cymene | Moringa Oleifera |
| 97 | Palmitoleic acid | Moringa Oleifera |
| 98 | Procyanidin | Moringa Oleifera |
| 99 | Pterygospermin | Moringa Oleifera |
| 100 | Quercetin | Moringa Oleifera |
| 101 | Rutin | Moringa Oleifera |
| 102 | Salicylic acid | Moringa Oleifera |
| 103 | Sinalbin | Moringa Oleifera |
| 104 | Sinapic acid | Moringa Oleifera |
| 105 | Squalene | Moringa Oleifera |
| 106 | Stearic acid | Moringa Oleifera |
| 107 | Syringic acid | Moringa Oleifera |
| 108 | Tetracontane-1,40-diol | Moringa Oleifera |
| 109 | Trimethyl (4-tert-butylphenoxy)silane | Moringa Oleifera |
| 110 | Vanillin | Moringa Oleifera |
| 111 | Vicenin-2 | Moringa Oleifera |
| 112 | Z,Z-2,5-Pentadecadien-1-ol | Moringa Oleifera |
| 113 | Ramonanin A | Lignum Vitae |
| 114 | Ramonanin B | Lignum Vitae |
| 115 | Ramonanin C | Lignum Vitae |
| 116 | Ramonanin D | Lignum Vitae |
| 117 | Gingerenone B | Lignum Vitae |
| 118 | Glucoputranjivin(1−) | Lignum Vitae |
| 119 | β-sesquiphellandrene | Lignum Vitae |
| 120 | Glucoputranjivin | Lignum Vitae |
| 121 | Pinocarveol | Lignum Vitae |
| 122 | (2E,6E)-hedycaryol | Lignum Vitae |
| 123 | Dodecane | Lignum Vitae |
| 124 | Zingiberene | Lignum Vitae |
| 125 | all-cis-octadeca-6,9,12,15-tetraenoic acid | Lignum Vitae |
Table 1: Complete list of compounds and their origin.
The virtual screening and molecular docking were performed using the Schrodinger small molecule drug discovery suite, Schrödinger Release 2020-2: Schrödinger, LLC, New York, NY, 2020.
Protein preparation and receptor grid generation
The COVID-19 proteins, main protease (PDB ID: 6LZE), RNA dependent RNA polymerase (PDB ID: 7BV2) and nsp-15 (PDB ID: 6WLC) were retrieved from the Protein Data Bank [12,13]. Using Schrodinger’s Protein Preparation Wizard the imported proteins were preprocessed, optimized, and minimized. During preprocessing of proteins missing hydrogens and residues were added with the help of Prime (version). As there was a bound ligand with all these proteins, so the receptor grids with the dimensions 10 Å × 10 Å × 10 Å were generated by selecting the atom of ligand molecule using Schrodinger receptor grid version 8.7.
Ligand preparation
A total of 4 medicinal plants native to the Caribbean were selected to compile a list of compounds to be virtually screened and identify potential antiviral agents for COVID-19. A literary analysis of numerous research papers and screening of databases generated compounds from each of the 4 plants for a total of 125 compounds (Table 1). The 2D chemical structures of all compounds discovered were imported from the PubChem database in SDF format into Maestro. Using Schrodinger’s LigPrep module, all the compounds were converted into 3D structures and optimized under Force field OPLS3e, different ionization states were generated at pH 7.0±2.0 using Epik module (version 5.2) Specific chiralities of 3D structures were retained, while around other chiral centers all possible stereoisomers were generated. Total 153 structures were taken to the docking analysis.
In present study, co-crystallized ligand of each protein was used as reference ligand; a peptide inhibitor {N}-[(2~{S})-3- cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})- 2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]- 1~{H}-indole-2-carboxamide (FHR) (Figure 1a) of main protease, Ramdesivir (Figure 1b) as an inhibitor of RdRp, and Uridine-5’- monophosphate (Figure 1c) inhibitor of nsp-15.
Figure 1a: {N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1- oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl] amino]propan-2-yl]-1~{H}-indole-2-carboxamide (FHR).
Figure 1b: Ramdesivir.
Figure 1c: Uridine-5’-monophosphate.
Molecular docking
Using Glide Standard Precision (SP) the interaction between each protein and the 153 compounds and native ligand were analyzed. Based on the binding affinity scores generated (docking score), the compounds that had scores close to or better than the native ligand were selected for Extra Precision (XP) docking. Of the 153 docked compounds those with docking scores better than the native ligand for the protein were selected for further screening in XP mode along with the native ligand. If no compounds had a better docking score than the native ligand, compounds with glide scores-6.00 were chosen and further screened. The docking scores were again analyzed to determine which compounds, if any, have a higher affinity than the native ligand.
Further analysis of the compounds was necessary to determine the binding energy of protein-ligand complexes. With the knowledge of which compounds and poses best interacted with the protein, the compounds from XP mode were analyzed using Prime MM- GBSA. The SP and XP docking results showed that the various compounds interact with the protein and bind to the active site while Prime-MM-GBSA shows the free binding energy of the ligand-protein association.
ADME properties
The conclusion of the molecular docking study identified different compounds that interact well with the protein in comparison to the respective native ligand. Using QikProp (v6.4) of the Maestro (v12.4), the ADME properties of the top docking scorers were analyzed to determine if any of the compounds showed drug-like properties.
Structure-based virtual screening
Using the binding site of the native ligand, FHR, bound to the main protease 6LZE, a receptor grid of the active site was generated to dock the researched 125 compounds. An analysis of the active site showed the native ligand interaction via hydrogen bonds (H-Bond) with the GLY 143, PHE 140, HIE 163, and GLU 166 residues (Figures 2a-2c). Ligand preparation of the 125 researched compounds generated a total of 153 different poses to be filtered with all 153 meetings the filter parameters. Because of the small compound library, preliminary virtual screening was carried out in SP mode and a total of 38 compounds were filtered out based on their docking score. The SP mode docking results did not yield any compounds with a docking score better than the native (docking score=-9.734) so all compounds with scores equal to or greater than-6.00 were selected for XP virtual screening.
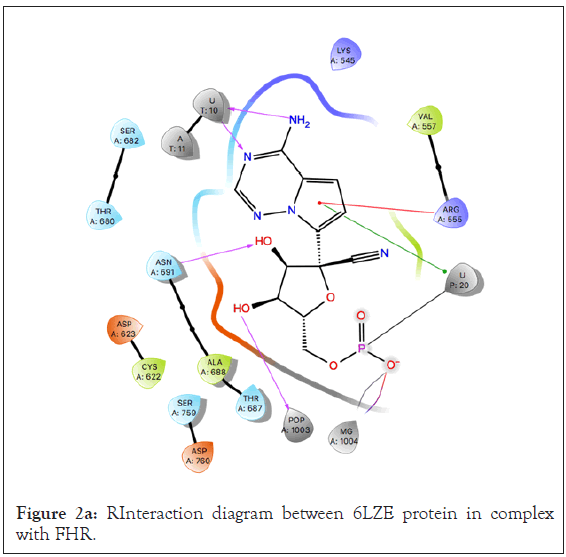
Figure 2a: RInteraction diagram between 6LZE protein in complex with FHR.
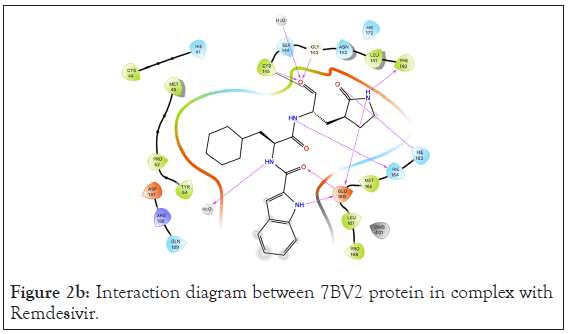
Figure 2b: Interaction diagram between 7BV2 protein in complex with Remdesivir.
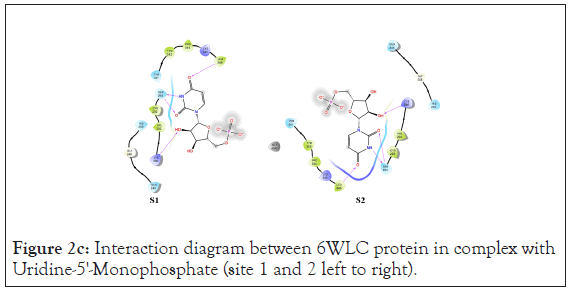
Figure 2c: Interaction diagram between 6WLC protein in complex with Uridine-5'-Monophosphate (site 1 and 2 left to right).
Like the 6LZE protein, 7BV2 and 6WLC had their active site analyzed and the compounds docked to those sites in SP mode to determine top scorers. There were 40 compounds that had better docking scores than the native ligand when docked in SP mode to 7BV2. On the other hand, because 6WLC had 2 different sites, the compounds were docked in SP mode to both. Site 1 did not produce any compounds that had better docking scores than the native so all compounds with SP scores >6.00 were selected for XP docking. Site 2 on the other hand had 18 compounds with better docking scores than the native in SP mode. All compounds that scored better than the native or had an SP docking score >6.00 when no better interactions were produced were docked to their respective sites in XP mode.
The XP docking method was able to provide positive results as it showed there were compounds that interacted better than the native ligand. The XP docking yielded multiple compounds that had a better docking score than the native ligand for each protein (Tables 2-4).
| Compounds | XP docking score | Molecular Weight | Prime MMGBSA dG Bind (kcal/mol) |
|---|---|---|---|
| Rutin | -11.962 | 610.524 | -70.54 |
| Vicenin-2 | -10.001 | 594.525 | -60.39 |
| Procyanidins | -9.747 | 594.528 | -56.41 |
| Kaempferol-3-Rutinoside | -9.388 | 594.525 | -47.29 |
| Astragalin | -9.369 | 448.382 | -42.34 |
| Kaempferol-3-O-Glucoside | -9.369 | 448.382 | -42.34 |
| 10-Hydroxyaloin A | -9.217 | 434.399 | -54.61 |
| Marumoside B | -9.031 | 459.449 | -23.83 |
| Multiflorin B | -9.045 | 594.525 | -22.96 |
| Orientin | -8.554 | 448.382 | -54.66 |
| Chlorogenic Acid | -8.373 | 354.313 | -18.06 |
| Kaempferol-3-O--Rhamnoside | -8.354 | 756.667 | -40.47 |
| Aloin | -8.233 | 418.399 | -52.31 |
| FHR | -8.095 | -69.71 |
Table 2: Top scoring compounds from 6LZE docking.
| Compounds | XP docking score | Molecular Weight | Prime MMGBSA dG Bind (kcal/mol) |
|---|---|---|---|
| Vicenin-2 | -8.194 | 594.525 | -44.54 |
| Multiflorin B | -7.657 | 594.525 | -29.57 |
| Marumoside B | -7.591 | 459.449 | -38.5 |
| 3,4-Di-O-Caffeoylquinic Acid | -7.585 | 516.457 | -39.88 |
| N-Alpha-L-Rhamnopyranosyl Vincosamide | -7.468 | 660.674 | -39.3 |
| Isoquercetin | -7.467 | 464.382 | -32.69 |
| Swertiajaponin | -7.245 | 462.409 | -29.12 |
| Orientin | -7.09 | 448.382 | -48.84 |
| Kaempferol-3-Rutinoside | -7.008 | 594.525 | -41.88 |
| Aloin | -6.912 | 418.399 | -41.95 |
| Glucotropeolin | -6.561 | 409.425 | -22.07 |
| Chlorogenic Acid | -6.412 | 354.313 | -22.96 |
| Astragalin | -6.337 | 448.382 | -32.72 |
| Kaempferol-3-O-Glucoside | -6.337 | 448.382 | -32.72 |
| D-Allose | -6.184 | 180.157 | -15.92 |
| Sinalbin | -6.18 | 425.425 | -28.97 |
| Niazimicin | -6.147 | 357.421 | -19.05 |
| Isoaloeresin D | -6.136 | 556.565 | -42.16 |
| Cryptochlorogenic Acid | -6.017 | 354.313 | -27.31 |
| Glucoconringiin | -6.004 | 391.408 | -20.93 |
| Glucoconringiin 1_ | -6.004 | 391.408 | -20.93 |
| Glucoputranjivin | -5.971 | 361.381 | -22.8 |
| Glucoputranjivin 1 | -5.971 | 361.381 | -22.79 |
| Isovitexin | -5.987 | 432.383 | -30.58 |
| 10-Hydroxyaloin A | -5.925 | 434.399 | -30.2 |
| Isorhamnetin | -5.645 | 316.267 | -9.1 |
| Aloesin | -5.617 | 394.377 | -34.93 |
| Kaempferol-3-O-Alpha-Rhamnoside | -5.406 | 756.667 | -37.95 |
| Moringyne | -5.286 | 312.319 | -20.33 |
| Niaziminin | -4.984 | 399.458 | -35.99 |
| Niazirin | -4.915 | 279.292 | -32.1 |
| Caffeic Acid | -4.848 | 180.16 | -22.94 |
| Aloe Emodin Anthrone | -4.815 | 256.257 | -16.68 |
| Gallic Acid | -4.798 | 170.121 | -18.32 |
| O-Ethyl-4-[(Alpha-L-Rhamnosyloxy)-Benzyl] Carbamate | -4.635 | 357.36 | -29.36 |
| Remdesivir (Native) | -4.566 | -44.66 |
Table 3: Top Scoring Compounds from 7BV2 docking.
| XP docking score | Molecular Weight | Prime MMGBSA dG Bind (kcal/mol) | |
|---|---|---|---|
| Orientin S1 | -10.896 | 448.382 | -10.24 |
| Uridine-5'-Monophosphate S1 | -9.035 | 0 | |
| Swertiajaponin S2 | -12.364 | 462.409 | -10.24 |
| Glucoconringiin S2 | -11.691 | 391.408 | 0 |
| Sinalbin S2 | -11.543 | 425.425 | 0 |
| Glucoconringiin 1- S2 | -11.153 | 391.408 | 0 |
| Aloesin S2 | -10.411 | 394.377 | -10.24 |
| Uridine-5'-Monophosphate S2 | -10.325 | 0 | |
| 6WLC has 2 binding sites. The corresponding site for the compound is denoted as S1 (site 1) and S2 (site 2) | -4.566 | -4.566 | -4.566 |
Table 4: Top scoring compounds from 6WLC docking.
The 6LZE protein had 13 compounds that docked better than the native ligand. These 13 interactions were analyzed using Prime MM-GBSA to determine the binding energy. Glide XP and Prime MM-GBSA scores for the top 13 compounds can be found in Table 2. The most prominent interaction between the protein- ligand complex was hydrogen bonding. In almost all complexes, hydrogen bonding occurred between the compound and the GLU 166 residue as is present in the native ligand-protein complex. This is the only common interaction when comparing the native ligand and the tested compounds, however when comparing the test compounds, hydrogen bonding with the CYS 145 residue and pi-pi stacking interaction between the HIE 41 residue and benzene derivatives are prominent (Figures 3a and 3b).
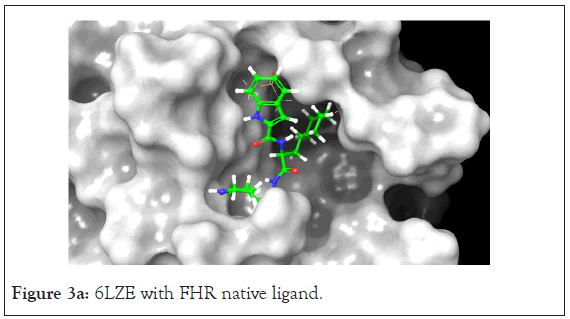
Figure 3a: 6LZE with FHR native ligand.
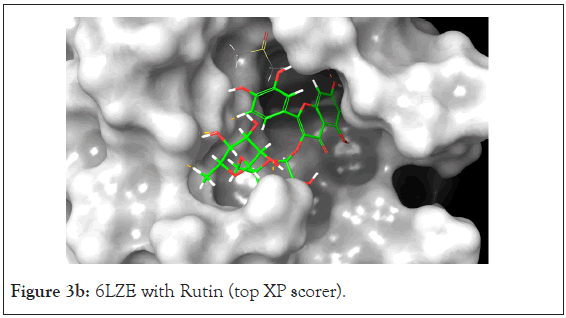
Figure 3b: 6LZE with Rutin (top XP scorer).
The Prime MM-GBSA analysis showed the free binding energy of each compound-protein complex. The top 13 compounds had a range of free binding energy from-18.06 kcal/mol to-70.54 kcal/ mol. The complex with the lowest energy, which is most favorable, belongs to Rutin which also has better binding energy than the native ligand which would suggest a potentially strong inhibitor of the main protease. Rutin also had the best docking score at-11.962. The ligand interaction diagram shows H-Bonds with GLY 143 and GLU 166 residues the same as the native ligand but also interacts via hydrogen bonding with ASN 142, CYS 145, HIE 41, and GLN 189.
A total of 36 compounds had better ligand interactions than the native ligand of 7BV2. Similar to 6LZE, the main interactions between the protein and the ligands is hydrogen bonding. The native ligand interacts with the protein by hydrogen bonds with ASN 691, U10, POP 1003 and ring interactions with U 20 and ARG 555. The binding energy for the top compounds range from- 9.1 to-48.84. The compound Orientin has the most favorable Peime-MM GBSA binding energy (-48.84) and has a better docking score than the 7BV2 native ligand. The ligand interaction diagram for Orientin shows that its interaction with the protein is strictly via hydrogen bonding with A 11, U 12, A 13, A 12, A 14, C 15, ASN 496 (Figures 4a and 4b).
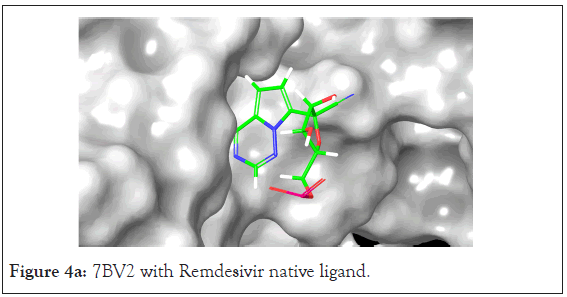
Figure 4a: 7BV2 with Remdesivir native ligand.
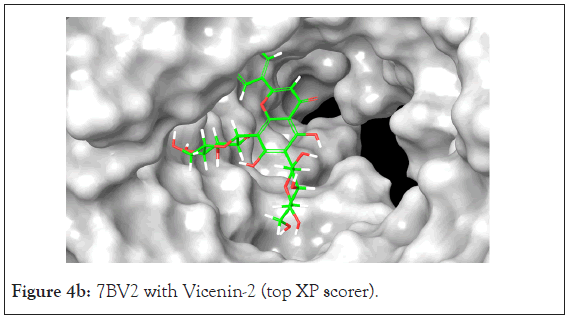
Figure 4b: 7BV2 with Vicenin-2 (top XP scorer).
An analysis of the docking results for the 6WLC protein shows that one site has more favorable interactions with the compounds than the others. Only 1 of the 125 compounds interacts more favorably at site 1 (S1) than the native. Site 2 (S2) on the other hand had favorable interactions with 5 compounds (Table 4). The native ligand, Uridine-5'-Monophosphate, interacted with the protein via hydrogen bonding with LYS 65, ILE 64, SER 294 and LEU 346. Comparison of the native ligand interaction and the tested compounds shows that there are many residues in common between them. The tested compounds all form hydrogen bonds with at least one of the same residues as the native suggesting their importance in the active site and compound interactions (Figures 5a and 5d).
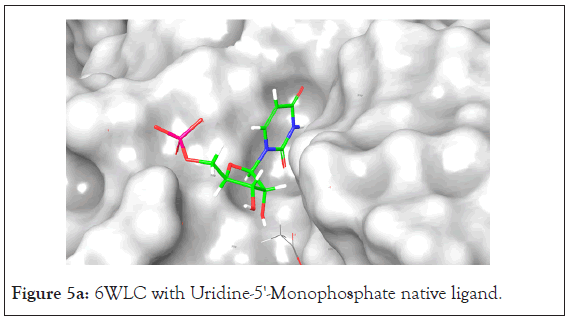
Figure 5a: 6WLC with Uridine-5'-Monophosphate native ligand.
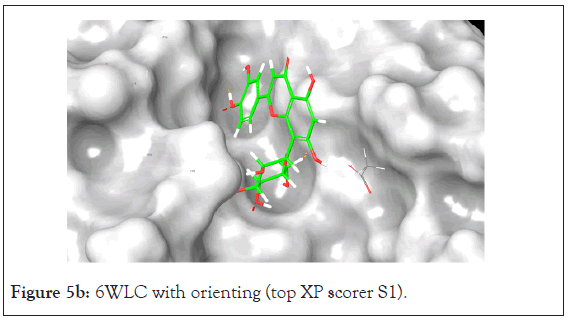
Figure 5b: 6WLC with orienting (top XP scorer S1).
In addition to the docking score, Prime MMGBSA analysis shows that these compounds have the most favorable binding energy out of all the protein-ligand interactions. The binding energy for the native ligand is 0 kcal/mol for both sites. Of the 6 total ligands that interacted with the protein irrespective of site, half have equal binding energy to the native and the other half have better binding energy than the native at-12.04 kcal/mol. The common residue interaction among all compounds aside from Aloesin is the SER 294 residue, suggesting it is a key residue in ligand interaction in the active site. A complete list of H-bonding interactions for the top scoring compounds is given in Tables 2-5.
| Compounds | Residues forming H-Bonding |
|---|---|
| Orientin | SER 294, GLN 245, THR 341, HIS 250, HIS 235 |
| Swertiajaponin | SER 294, LYS 290, LYS 174, GLU 146, LEU 346, ASP 17 |
| Glucoconringiin | TYR 343, SER 294 |
| Sinbalin | SER 294, VAL 292 |
| Glucoconringiin 1 | GLU 146, LYS 174, SER 294, LYS 65 |
| Aloesin | HIS 235, GLN 245, GLU 146, LYS 65 |
Table 5: H-bonding interactions of Top Scoring Compounds with different residues of 6WLC.
Among all the compounds, one showed promising interaction with all three proteins. Orientin, a compound native to Aloe Vera, not only showed docking scores that were better than the native ligand for all proteins but also had a better binding affinity to the protein than the native. It showed higher affinity with the 7BV2 and 6WLC proteins and had a similar score for 6LZE. Prime MMGBSA and XP docking scores for Orientin and each protein’s native ligands can be found in their respective Tables 2-4 for 6LZE, 7BV2 and 6WLC respectively Table 6.
| Compound | Rule of 5 | Donor HB | Accept HB | % Human oral absorption | QPlogS | QPlogPo/w | QPlogKhsa | #metab | QPlogHERG | QPPCaco | QPlogBB |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Rutin | 3 | 9 | 20.55 | 0 | -2.114 | -2.536 | -1.293 | 10 | -5.217 | 1.158 | -4.419 |
| Vicenin-2 | 3 | 10 | 20.75 | 0 | -3.016 | -2.851 | -1.333 | 15 | -6.03 | 0.498 | -5.182 |
| Procyanidins | 3 | 10 | 11.65 | 0 | -3.622 | 0.082 | -0.394 | 12 | -5.892 | 1.024 | -4.313 |
| Kaempferol-3-rutinoside | 3 | 8 | 19.8 | 0 | -3.129 | -1.944 | -1.281 | 9 | -6.392 | 1.458 | -4.813 |
| Astragalin | 2 | 6 | 13 | 11.182 | -2.741 | -0.822 | -0.79 | 7 | -5.422 | 6.86 | -3.27 |
| Kaempferol-3-O-glucoside | 2 | 6 | 13 | 11.182 | -2.741 | -0.822 | -0.79 | 7 | -5.422 | 6.86 | -3.27 |
| 10-Hydroxyaloin A | 1 | 6 | 12.45 | 22.877 | -2.58 | -0.895 | -0.753 | 11 | -4.86 | 6.16 | -3.219 |
| Marumoside B | 2 | 8 | 18.55 | 0 | -0.849 | -3.009 | -1.674 | 8 | -3.63 | 4.509 | -3.442 |
| Multiflorin B | 3 | 8 | 19.8 | 0 | -2.513 | -2.031 | -1.225 | 9 | -5.74 | 1.681 | -4.334 |
| Orientin | 2 | 7 | 13 | 5.071 | -2.752 | -1.178 | -0.747 | 10 | -5.024 | 4.084 | -3.343 |
| Chlorogenic acid | 1 | 6 | 9.65 | 17.723 | -2.532 | -0.239 | -0.932 | 5 | -3.304 | 1.936 | -3.291 |
| Kaempferol-3-O--rhamnoside | 3 | 11 | 28.3 | 0 | -2.411 | -4.255 | -2.13 | 12 | -6.635 | 0.09 | -7.139 |
| Aloin | 1 | 5 | 11.7 | 30.587 | -2.657 | -0.392 | -0.623 | 11 | -4.693 | 11.367 | -2.803 |
| 3,4-Di-O-caffeoylquinic acid | 3 | 7 | 11.45 | 1 | -4.307 | 0.812 | -0.628 | 6 | -4.935 | 0.199 | -5.305 |
| N-alpha-L-rhamnopyranosyl vincosamide | 3 | 7 | 23.9 | 1 | -2.218 | -1.998 | -1.346 | 11 | -6.917 | 1.673 | -3.594 |
| isoquercetin | 2 | 7 | 13.75 | 1 | -2.68 | -1.394 | -0.874 | 8 | -5.379 | 2.192 | -3.894 |
| swertiajaponin | 2 | 6 | 13 | 1 | -3.292 | -0.531 | -0.696 | 10 | -5.544 | 9.208 | -3.205 |
| Glucotropeolin | 0 | 5 | 14 | 1 | -1.37 | -1.098 | -1.339 | 6 | -2.64 | 2.314 | -3.061 |
| D-allose | 0 | 5 | 10.2 | 2 | -1.045 | -2.211 | -0.873 | 4 | -2.7 | 67.286 | -1.569 |
| Sinalbin | 2 | 6 | 14.75 | 1 | -1.404 | -1.559 | -1.396 | 7 | -2.608 | 0.954 | -3.515 |
| niazimicin | 0 | 4 | 9.55 | 3 | -3.631 | 1.179 | -0.483 | 4 | -5.179 | 264.212 | -1.521 |
| Isoaloeresin D | 2 | 5 | 14.5 | 1 | -3.783 | 1.588 | -0.342 | 10 | -5.402 | 59.493 | -2.531 |
| Cryptochlorogenic acid | 1 | 6 | 9.65 | 2 | -2.115 | -0.179 | -0.913 | 5 | -2.774 | 3.012 | -2.857 |
| glucoconringiin | 2 | 6 | 14.75 | 1 | -1.458 | -1.837 | -1.511 | 7 | -2.598 | 1.314 | -3.661 |
| glucoconringiin 1- | 2 | 6 | 14.75 | 1 | -1.458 | -1.837 | -1.511 | 7 | -2.598 | 1.314 | -3.661 |
| Glucoputranjivin | 0 | 5 | 14 | 2 | -1.392 | -1.477 | -1.412 | 6 | -2.241 | 3.359 | -2.961 |
| Glucoputranjivin 1 | 0 | 5 | 14 | 2 | -1.392 | -1.477 | -1.412 | 6 | -2.241 | 3.359 | -2.961 |
| Isovitexin | 1 | 6 | 12.25 | 2 | -3.29 | -0.591 | -0.671 | 9 | -5.688 | 8.105 | -3.185 |
| isorhamnetin | 0 | 3 | 5.25 | 3 | -3.458 | 1.232 | -0.152 | 5 | -5.169 | 59.672 | -1.952 |
| Aloesin | 0 | 5 | 13.75 | 2 | -2.747 | -1.03 | -0.884 | 9 | -4.726 | 22.527 | -2.648 |
| Moringyne | 0 | 4 | 10.5 | 3 | -2.22 | -0.087 | -0.767 | 6 | -4.43 | 212.456 | -1.449 |
| niaziminin | 0 | 3 | 9.85 | 3 | -5.202 | 2.314 | -0.207 | 3 | -5.984 | 387.686 | -1.461 |
| Niazirin | 0 | 3 | 9.05 | 3 | -3.062 | -0.003 | -0.75 | 4 | -4.469 | 145.234 | -1.608 |
| Caffeic acid | 0 | 3 | 3.5 | 2 | -1.369 | 0.562 | -0.792 | 2 | -2.197 | 21.699 | -1.576 |
| Aloe emodin anthrone | 0 | 1 | 3.2 | 3 | -3.205 | 2.016 | 0.027 | 4 | -4.568 | 224.785 | -1.188 |
| Gallic acid | 0 | 4 | 4.25 | 2 | -0.701 | -0.567 | -0.983 | 3 | -1.417 | 10.04 | -1.662 |
| O-ethyl-4-[(alpha-L-rhamnosyloxy)-benzyl] carbamate | 0 | 5 | 8.6 | 3 | -3.142 | 0.681 | -0.468 | 4 | -4.952 | 65.672 | -2.232 |
Table 6: ADME Properties.
ADME properties
The 13 best compounds from molecular docking went through ADME analysis using the QikProp module on Maestro. A total of 11 properties were the main focus which include the molecular weight, the number of hydrogen donors, the number of hydrogen acceptors, the number of violations of the rule of five, predicted IC50 value for blockage of HERG K+ channels (QPlogHERG), percent human oral absorption, predicted aqueous solubility (QPlogS), prediction of binding to human serum albumin (QPlogKhsa), predicted octanol/water partition coefficient (QPlogPo/w), predicted blood/brain partition coefficient (QPlogBB), number of likely metabolic reactions (#metab), and predicted apparent Caco-2 cell permeability (QPPCaco-).
ADME analysis showed that each compound has its pros and cons with no one compound falling within the acceptable range for all the properties. Most of the compounds had at least 1 violation of the rule of five with a few that had no violations. Lipinski Rule of Five states that drug-like compounds should have molecular weight lower than 500, lipophilicity (logP) lower than 5, less than five hydrogen bond donors, and less than 10 hydrogen bond acceptors, but many of natural products drugs do not comply with the “Rule of Five” [14]. it is recommended to not apply overly rigid cut-off points, as it increases the risk of losing some valuable compounds in earlier stages of screening [14], especially in case of natural products. A comprehensive list of the compounds and their ADME properties can be found in Table 6.
The researched plants are all used for medicinal purposes in the Caribbean from centuries. Based on the present in-silco study, multiple compounds showed promising inhibitory potential against different SARS CoV-2 proteins. Some of the compounds like rutin, vicenin-2, and Orientin exhibited better protein- compound association in compare to respective native ligands and found to have more negative binding energy. Further research on the antiviral properties of these plants against the various proteins of the SARS CoV-2 virus could prove useful for the discovery of naturally occurring antiviral compounds for future drugs and may provide natural remedies against this fatal disease.
Citation: Duncan A, Margetson J, Roberts J, Baron T, Buxani N (2021) Caribbean Plants as Source of Novel Inhibitors for Main Protease, NSP-15, and RNA-dependent RNA polymerase (RdRp) of SARS-Cov-2. Drug Des. 10:173.
Received: 21-Dec-2020 Accepted: 04-Jan-2021 Published: 11-Jan-2021 , DOI: 10.35248/2169-0138.21.10.173
Copyright: © 2021 Duncan A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.