Journal of Leukemia
Open Access
ISSN: 2329-6917
ISSN: 2329-6917
Research Article - (2019)Volume 7, Issue 1
We investigated the infection of Bovine Leukemia Virus (BLV) related to other pathogens [Neospora caninum, Bovine Herpesvirus-1 (BoHV-1), Bovine Viral Diarrhea Virus (BVDV), and pathogenic bacteria] in 80 bovine aborted fetuses. The materials comprised whole fetuses, fetal organs, and placenta. The BLV was diagnosed by nested-PCR (env gp51 BLV gene), the identification of viral genotypes by sequencing, and the phylogenetic analysis by neighborjoining and maximum composite likelihood methods. The other pathogens and diagnoses were, respectively: Neospora caninum (nested-PCR), BoHV-1 (nested-PCR), BVDV (PCR), Brucella spp. (isolation and identification), Leptospira spp. (PCR), aerobic bacteria [Enterobacteriaceae, Gram positive cocci, Trueperella (Arcanobacterium) pyogenes] and micro-aerophilic (Campylobacter spp., Histophilus somni, and Listeria monocytogenes) by isolation and identification. BLV fetal antibodies were identified by ELISA kit. Thirteen (16.25%) fetuses were positive by BLV nested-PCR. Phylogenetic analysis revealed BLV genotypes 1, 5, and 6, which are frequently found in cattle in Brazil, Argentina, and Uruguay. No fetuses were positive for BLV antibodies by ELISA. A single case of coinfection with BLV was found for each of the pathogens Trueperella (Arcanobacterium) pyogenes, Klebsiella spp., and Streptococcus spp. were isolated as a pure or representing the preponderance of bacteria in a pooled culture. In the 67 BLV-negative fetuses, pathogens identified were single cases of Trueperella (Arcanobacterium) pyogenes, Staphylococcus aureus, and Brucella abortus; 2 of Escherichia coli; 3 of bovine viral diarrhea virus; and 4 of Neospora caninum. No pathogens were found in 55 fetuses. The low number of BLV positive samples infected or no by other pathogens didn´t allow performing statistical analysis for understanding if there were significative differences among not infected and infected BLV fetuses. Because BLV is an immunosuppressive agent and predisposes the cow to other pathogens, its connection with Leukemia or abortions need additional studies with bigger sampling, for elucidating pathogenesis in the pregnant cow and in the fetus. The rates of BLV transplacental transmission show the necessity of prophylactic measures in Brazilian cattle herds, in order to avoid infection in utero.
Abortion; Bovine leukemia virus; Cattle; Fetuses; Filogeny; Histopathology; Nested PCR
Abortion and neonatal mortality are frequent causes of economic loss in bovine production [1]. Infectious agents such as viruses, bacteria, and parasites can be involved [2-4]. It was reported that of 2,544 cases of bovine abortion, pathogens were found in only 30.2% [5]. In the majority, it is not possible to define the etiology, due to autolysis, multiple potential causes, and insufficient sampling of fetal tissues, placenta, and maternal and fetal serum [6]. Worldwide fetal death in cattle is estimated to be 5% [7], but abortion outbreaks, extended calving interval, loss of milk production, and lower pregnancy rates have been cited [8].
In Brazil were reported several infectious agents causing bovine abortion [9-12], but in 50% to 70% of cases direct diagnosis was not possible. Mononuclear inflammatory infiltrate has been observed in high rates in aborted fetal tissue, suggesting infectious causes [1,13]. Thus the improvement in sensitivity and specificity of laboratory techniques to investigate prevalent pathogens as well as other infectious agents not currently considered in a program of sanitary surveillance for abortion is important [2].
Bovine Leukemia Virus (BLV), a delta retrovirus in the family Retroviridae, is the causative agent of bovine leukemia [14], a chronic disease in which seropositive animals are carriers [15]. Thirty to 70% can present persistent lymphocytosis, and, of these, 2% to 5% may develop lymphosarcoma [15,16].
This disease reaches high rates of seroreactivity in intensively managed dairy cattle, and has been reported in all regions of Brazil [17]. There is no specific legislation for BLV control in the country, although it is recommended by the World Organization for Animal Health that countries importing live animals, semen, and embryos should certify them as BLV-free [18]. The high prevalence and wide distribution of BLV in Brazil suggests that the virus should be investigated as a potential pathogen involved in bovine abortion. A primary characteristic of BLV is mononuclear inflammatory infiltrate in multiple organs of immature and adult cattle [15,19] and this type of cell has been observed in aborted bovine fetuses without conclusive differential diagnosis, which may indicate an infectious etiology [1,9,13].
The intrauterine (vertical) transmission of BLV occurs in the first semester of gestation in up to 8% of seropositive pregnant bovines, mainly in those with high virus concentration and low antibody titers [20]. Serology of newborn calves before they ingest colostrum has been used to diagnose vertical BLV transmission. Two reports revealed 20% [21] and 4.8% [22], of neonates seropositive by Agar Gel Immunodiffusion test (AGID), and another study [23] detected in two herds 10% and 13% of neonates seropositive by AGID, with 15% and 22% confirmed by ELISA. Sacrificed BLV infected pregnant cows had 33.3% of 5-9 month fetuses seropositive to BLV by AGID [24].
Since bovine leukemia is prevalent in Brazilian cattle, and BLV in aborted bovine fetuses has not been investigated, the aim of this research was to investigate the infection of BLV in aborted fetuses using molecular techniques (nested-PCR, DNA sequencing, and phylogeny), ELISA, and examination of histological lesions suggestive of viral abortion, as well as examination of BLV positive fetuses for co-infection with other pathogens.
This research was approved by the Bioethics Committee in Animal Research of the Biological Institute (CETEA-IB), registry number 122/12 according the Sociedade Brasileira de Ciência em Animais de Laboratório/Colégio Brasileiro de Experimentação Animal (SBCAL/COBEA).
Eighty bovine abortion cases from several Brazilian states were examined. Material comprised whole fetuses, fetal organs, and placentas. Materials were frozen or refrigerated and transported to the Centro de Pesquisa de Sanidade Animal do Instituto Biológico (CPSA-IB) from December, 2007 to October, 2012. When necropsy was performed by veterinarians in the field, the organs sampled varied, so sample numbers differed among tissues. The necropsies performed in an appropriate facility at CPSA-IB used sterilized instruments and the materials collected (organs, abomasal contents, thoraco-abdominal fluid) were placed in sterile bottles according to described protocols [25].
Estimates of the gestational age of fetuses were determined by one of three methods: crown-rump length [26], breeding dates, or estimates made by veterinarians.
Histology was performed on samples of thymus, spleen, lymph node, lung, heart, liver, kidney, adrenal gland, and brain fixed in 10% buffered formalin. Tissues were cut into small pieces, dehydrated, cleared, and embedded in paraffin; they were cut into the microtome (three μm) and stained with hematoxylin and eosin [27].
Thymus, spleen, lymph node, placenta, and thoraco-abdominal fluid were submitted to nested-PCR for BLV pro-viral DNA analysis. The DNA was extracted from chilled tissue samples using the commercial kit DNeasy Blood & Tissue Qiagen (Valencia, CA, USA). Amplification of the segment that encodes the env gp51 BLV gene used specific external primers for amplifying a segment of 598 base pairs (bp) (BLV1–5’ TCT GTG CCA AGT CTC CCA GAT A 3’ and BLV2– 5’ AAC AAC AAC CTC TGG GAA GGG T 3’) and specific internal primers for amplifying a segment of 444 bp (BLV3– 5’ CCC ACA AGG GCG GCG CCG GTT T 3’ and BLV4–5’ GCG AGG CCG GGT CCA GAG CTG G 3’) [28]. The positive control was a continuous lineage cell (fetal lamb kidney) infected with BLV and the negative control was VERO cells and ultra-pure water. Each sample was incubated in the thermocycler Mastercycler (Eppendorf, Foster City, CA, USA). The first amplification used 20 μL of GoTaq Green Master Mix (Cat #M7502, Promega, Madison, WI, USA) with 6.5 μL nucleasefree water, 12.5 μL PCR buffer, 0.5 μL primer VLB1, and 0.5 μL primer VLB2 for 5 μL of DNA. The PCR conditions of the first amplification were initial denaturation at 94°C for 2 min; 40 repeat cycles of denaturation at 95°C for 30 sec; annealing at 62°C for 30 sec, extension at 72°C for 1 min, and a final extension at 72°C for 4 min. The second amplification used 22 μL of GoTaq Green Master Mix (Cat #M7122, Promega, Madison, WI, USA) with 9.0 μL nuclease-free water, 12.5 μL PCR buffer, 0.25 μL primer VLB3, 0.25 μL primer VLB4, and 3 μL of the final product of the first amplification, and each sample was incubated in the thermocycler. The conditions for the second amplification were denaturation at 94°C for 2 min, 40 cycles at 95°C for 30 sec, annealing at 70°C for 30 sec, extension at 72°C for 1 min, and a final extension at 72°C for 4 min. The analysis of amplified products was performed by electrophoresis (100 V/60 min) in a low melting point agarose gel (Agarosis Cat. #V31125, LE, Promega, Madison, WI, USA) at 1.5% in Buffer TAE (Cat. #V4281, Promega, Madison, WI, USA) pH 8.2-8.4 1x. The DNA samples were stained with 1 μL Gel Red nucleic acid stain (Biotium Inc., Hayward, CA, USA) diluted in ultra-pure water at 1:150 and 2 μL of loading (6X DNA Loading Dye, Thermo Scientific, Waltham, MA, USA). A 100 bp DNA ladder (Fermentas, Vilnius, Lithuania) was used. The gel image under UV light (320 nm) was recorded using a system for photodocumentation and analysis (Alpha Innotech, San Leandro, CA, USA).
Eight samples yielded sufficient DNA for sequencing, they were purified with the Qiaquick PCR Purification kit 250 (cat. #28106, Qiagen, Valencia, CA, USA) and subjected to sequencing by chain termination with dideoxynucleotides marked with fluorophores. The reaction was performed with 50 μL of the PCR product, 4 μL of each primer used in PCR (final concentration 3.2 μM) for a total volume of 10 μL, 3 μL of sequencing buffer 5 (5X concentration), and 1 μL of Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). Each sample was sequenced in both directions using forward and reverse primers. Each sample was incubated in the thermocycler for 35 cycles at 95°C for 10 sec and 60°C for 4 min. The sequencing reaction products were precipitated by adding 40 μL of 75% isopropanol and centrifuged at 3220 x g for 40 min at room temperature (Refrigerated Centrifuge UniCen MR, Herolab, Wiesloch, BW and Germany). The supernatant was discarded, and the plate was folded in absorbent paper and centrifuged at 3220 x g for 90 sec to remove residual isopropanol. The precipitate was washed with 100 μL of isopropanol 75%, and the supernatant was discarded without centrifugation. The precipitate was dried at 37°C for 10 min, re-suspended in 10 μL Hi-DI formamide (Applied Biosystems, Warrington, Chesire, UK), denatured at 95°C for 2 min, and subjected to capillary electrophoresis in a sequencer (3500 XL Genetic Analyzer, Applied Biosystems, Foster City, CA, USA).
The sequences were analyzed with Bioedit v.7.0.9 to generate a unique sequence from the bidirectional sequence data [29]. Sequences were aligned to one another, and to homologues sequences obtained from GenBank, using ClustalW version 1.8.3 software [29]. Phylogenetic inference was performed using the program Mega, v. 5.0 [30].
The BLV sequences recovered from Brazilian strains and from GenBank were used for analysis of genealogy. The access number, country of origin, and genotypes [31-35] were included for the determination of nucleotide identity using Bioedit v.7.0.9 software [29]. Construction of the phylogenetic tree used the method of maximum composite likelihood for the partial region (437 nt) of the env gp51 gene of BLV, with the bootstrap values from 1000 replicates, in MEGA v. 5.0 software [30].
Fetal immunodiagnosis was made by analysis of thoraco-abdominal fluid using ELISA kit for antibody detection of Bovine Leukemia Virus (Chekit Leucose serum antibody test kit, IDEXX Laboratories, Westbrook, USA), and the plates were read in an ELISA reader (Multiskan Ascent 354 Microplate Photometer, Thermo Scientific/ Labsystems, Waltham, MA, USA).
The bacteriological investigation considered microorganisms found in pure isolation or representing the preponderance of bacteria in a culture of pooled organs, (spleen, liver, lung, kidney), placenta, and abomasal contents. Brucella spp. was diagnosed by isolation and identification [36,37], and by PCR [38]. Leptospira spp. was diagnosed by PCR [38]. The isolation and identification of aerobic bacteria (Enterobacteriaceae, Gram positive cocci, Trueperella (Arcanobacterium) pyogenes) and micro-aerofilic (Campylobacter spp., Histophilus somni, and Listeria monocytogenes) used recognized protocols [6,12,37,39] Neospora caninum was diagnosed by nested- PCR [13], bovine herpesvirus-1 (BoHV-1) by nested-PCR [40], and Bovine Diarrhoea Virus (BDV) by PCR [41].
Nested PCR confirmed the presence of pro-viral DNA, showing BLV infection in 13 of the 80 fetuses (16.25%). This nested-PCR rate is similar to the seropositivity rates (AGID test) in newborn calves before colostrum ingestion [21-23,42]. The World Organization for Animal Health recommends the use of maternal and fetal blood serum for detection of BLV antibodies, and the fetal thoracoabdominal fluid for assessment of BLV pro-viral DNA by nested-PCR [37]. No sample of fetal thoraco-abdominal fluid was positive with ELISA, but two samples were nested PCR positive (Table 1). It was not possible to correlate results of indirect ELISA with those of nested PCR, since no fetus showed BLV antibodies with ELISA, indicating the importance of direct diagnosis of the pathogen using molecular techniques, which have greater sensitivity, since they detect animals in which BLV is in incubation or latency and that have not yet seroconverted [42-45].
Table 1: Distribution of 20 BLV-positive samples among lymphatic organs and thoraco-abdominal fluid identified by nested PCR, n=80 fetuses.
| Tissue and fluid samples | Number of cases | Number of positive tissues and fluids |
|---|---|---|
| Thoraco-abdominal fluid | 58 | 2 |
| Lymph node | 56 | 3 |
| Spleen | 76 | 7 |
| Thymus | 76 | 8 |
BLV pro-viral DNA is most readily identified in the lymphatic system, tissues recommended by World Animal Health Organization for detection [37]. In the present study, nested-PCR identified BLV in 8 thymus, 7 spleen, and 3 lymph node samples, and thymus and spleen were the most frequently sampled lymphatic organs, each at 76 (Table 1). Most positive fetuses showed BLV in a single tissue: thymus in 4, followed by spleen in 3, and in thoraco-abdominal fluid in two. However, four fetuses tested positive in more than one tissue-type (Table 2). Thus the diagnosis of BLV increases with examination of multiple organs. It is known that antigens administered to a fetus intravenously are captured in liver, spleen, and bone marrow and the immune response will occur primarily in spleen and lymph nodes [46].
Table 2: Cases of multiple organ BLV-positive fetuses identified with nested PCR, n=13.
| Infection combinations in single and multiple organs | Number of cases |
|---|---|
| Thymus | 4 |
| Spleen | 3 |
| Thoraco-abdominal fluid+spleen | 2 |
| Thoraco-abdominal fluid+spleen+thymus | 1 |
| Lymph node+thymus | 1 |
| Spleen+thymus | 1 |
| Lymph node + thymus | 1 |
| Thoraco-abdominal fluid | 0 |
| Lymph node | 0 |
| Thoraco-abdominal fluid+lymph node | 0 |
| Thoraco-abdominal fluid+thymus | 0 |
| Thoraco-abdominal fluid+lymph node+spleen+thymus | 0 |
| Lymph node+spleen | 0 |
| Lymph node+spleen+thymus | 0 |
Positivity for BLV by nested-PCR was detected at 3 to 6 months gestation in 5 of 13 aborted fetuses and in 7 of 13 at 6 to 9 months. In one positive fetus the gestation was not determined. A study found in 5 of 15 seropositive fetuses (5 to 9 months) from cows with advanced lymphosarcoma [24]. This was corroborated by the present finding of vertical transmission of BLV from the second trimester. BLV intrauterine transmission in the first trimester of pregnancy was reported in up to 8% of seropositive cows, mainly in those with a high concentration of virus and low antibody titers [20]. This indicates that the fetus can be infected in-utero, since in bi-ungulates the sindesmocorial placenta is impermeable to maternal immunoglobulins, and bovine fetuses are unable to produce antibodies before 100 days. Immunoglobulins, when found in fetuses, are produced by their own immune system [46].
The majority of BLV positive fetuses were 3+ months into gestation, and no thoraco-abdominal fluid was positive for BLV antibodies. This may have been due to a) Viral titer level insufficient to provoke an immune response, according to a report where no antibody production was observed in BLV infection of less than 100 copies of pro-viral DNA/μg [47]; b) A deficient primary immune response in which the immune system does not recognize the viral antigen, since BLV may decrease IgM producing cells in lymph node and spleen [48]; c) The long period required for BLV incubation to seroconvert [15]; and d) Viral latency due a plasma blocking factor (PBF) [49,50].
The sampling (80 cases) took place mainly in São Paulo with 39 fetuses and Minas Gerais State with 24. The BLV infection rate in São Paulo was 7 of 39 and, in Minas Gerais, 4 of 24, with a single positive case in the 5 samples from Goiás and in the 3 from Bahia. The highest frequency of fetuses BLV positive by nested-PCR was observed in dairy cattle, 8 of 13, in agreement with other studies [17].
Sequencing was possible in only 8 of the 13 samples positive for BLV because of the low amount of pro-viral DNA. This may be related to loss of DNA due to fetal autolysis, improper collection, preservation, and transportation of samples from the farm to the laboratory, or the unsatisfactory purification of the products sequenced. The 8 sequences had approximately 356 bp. The partial nucleotide alignment (356 nt) for the env gp51 gene of these samples: LAP 54363B (B, spleen), LAP 55150T (T, thymus), LAP 55704L (L, lymph node), LAP 56089T, LAP56089L, LAP 57020T, LAP 57474B and LAP 57095PL (P, placenta) was compared to Genbank sequences and other samples (VLB 05, 06, 07, 08, 09, 10, and 11) previously identified at the Instituto Biológico (IB) (unpublished data) (Table 3). Different clusters were verified in Brazilian samples. The phylogenetic analysis revealed 3 genotypes 1, 5, and 6. The sample LAP 56089L presented 100% nucleotide similarity with the sequence FJ808576 from Argentina [35] and the Brazilian JN254640 and JN254636 genotype 1 [33].
Table 3: Sequences with maximum and minimum similarity of nucleotides in the consensus region, when compared with one another, with GenBank, and other Instituto Biológico isolated sequences.
| Similarity | Nucleotide identity | |
|---|---|---|
| Maximum (100%) | Minimum (95.2%) | |
| Among the sequences of this study | LAP 54363B, LAP 56089T and LAP 56089L | LAP 54363B and LAP 55704B |
| LAP 57474B and LAP 55704 L | ||
| LAP 55704L, LAP54363B and LAP 56089L | ||
| Maximum (100%) | Minimum (95.2%) | |
| Among the sequences of the study, and with those from GenBank, and other Instituto Biológico samples | LAP 54363B and JN 254634 | LAP 54363 B and FJ 808578, FM 209475 and FM 209469 |
| LAP 55150T and JN 254639 | LAP 56089T and FJ 808578, FM 209475 and FM 209469 | |
| LAP 56089T and JN 254634 | LAP 57474B and FJ 808578, FM 209475 and FM 209469, JN 254637, JN 254635, JN 254633, JN 254638 and FJ 808582 | |
| LAP 56089L and VLB5, VLB 9, VLB 10, VLB 11, VLB 7, FJ 808576, JN 254640 and JN 254636 | LAP 55704 L and JN 254634 | |
| LAP 55704 and FJ 808578, FM 209469, FM 209475 | ||
L=Lymph node, T=Thymus, B=spleen, LAP=Laboratory of Anatomopathology of Instituto Biológico.
A phylogenetic tree (Figure 1) was constructed using the sequences of 356 (nt) from the env gp51 gene identified in the present study, sequences with major similarity recovered from GenBank, disposing access number, country of origin, and genotype [31-35], and other Brazilian strains VLB 05 to VLB 11 (Laboratório de Vroses de Bovídeos-Instituto Biológico-São Paulo-Brazil). The values of 1000 bootstrap replicates used the method of maximum composite likelihood.
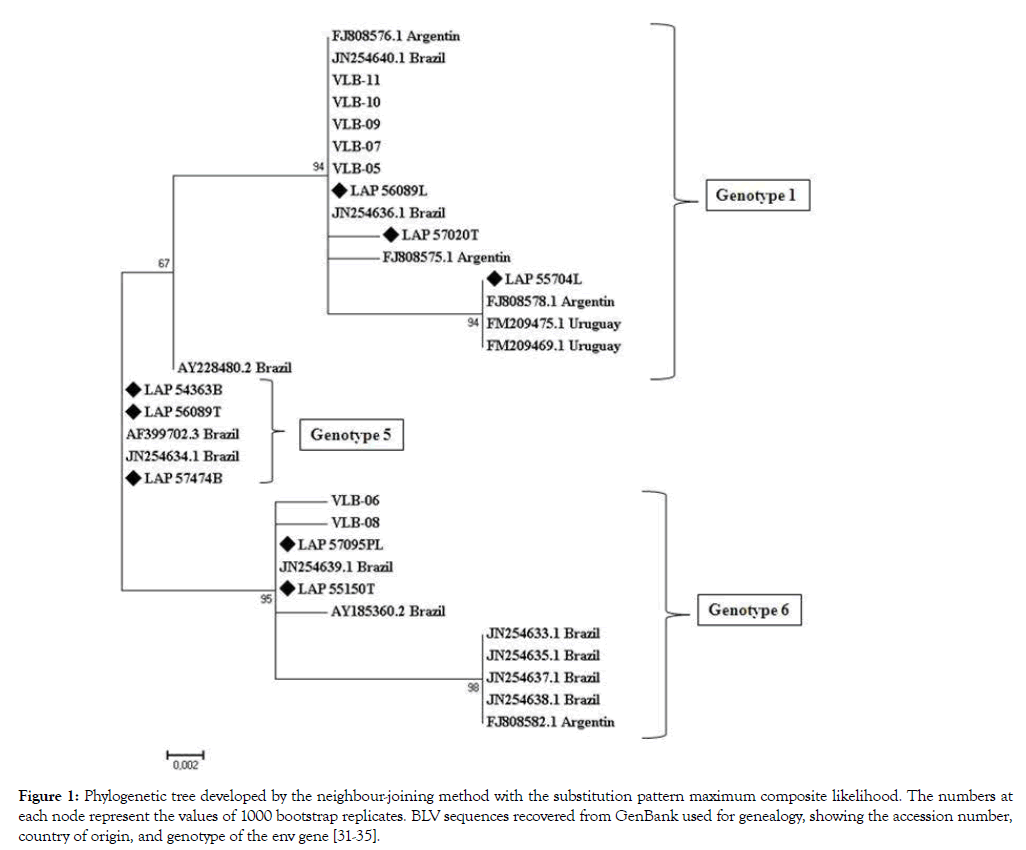
Figure 1: Phylogenetic tree developed by the neighbour-joining method with the substitution pattern maximum composite likelihood. The numbers at each node represent the values of 1000 bootstrap replicates. BLV sequences recovered from GenBank used for genealogy, showing the accession number, country of origin, and genotype of the env gene [31-35].
It is important to perform BLV molecular study in hosts, because the virus exhibits a diversity of genotypes characterized by similar sequences, but separated by longer branches [35]. The gp51 epitopes have high genetic variability that makes identification difficult. Several authors state that the molecular variations are responsible for differences between molecular and serologic diagnoses [51-53]. The sequence LAP 54363B/SP presented similarity with JN254634 found in São Paulo State (SP) [33], and belongs to genotype 5. The sample LAP 57474B from Minas Gerais State, when compared with the same cluster of the sample AF399702 from Minas Gerais [31] was also identified as genotype 5. Another study concluded that there was no correlation among sequences of Brazilian samples and viral pathogenicity and tissue tropism [32]. The present study suggests further research to investigate the association between BLV genotypes and clinical symptoms. Another study showed that, the sequence JN254639, genotype 6, has 100% similarity with LAP 55150T, both from São Paulo State [33]. Sample LAP 55704L is in the cluster of the sequences FM 209475 and FM 209469 (Uruguay) [34] and FJ808578 (Argentina) [35] with high bootstrap value (94) and belongs to genotype 1 (Figure 1, Table 3).
A previous report [34] identified samples in Brazil as 1, 5, 6, and 7, as in the present study, and the Uruguay samples only as genotype 1, revealing specific geographic BLV clustering. Sequence LAP 56089T showed maximum similarity to the Brazilian sample JN254634 genotype 5, and samples LAP 57095PL and 551570T presented a bootstrap value 95% in the cluster of the Brazilian sample JN254634, genotype 6 (Figure 1, Table 3).
The histological changes observed in BLV-positive fetuses were hyperplasia of thymus, spleen, and lymph node; and mononuclear inflammatory infiltrate in brain, lung, heart, liver, kidney, adrenal gland, and placenta, characterizing a generalized lymphoproliferative reaction and suggesting a cellular reaction to an infectious agent. Infection by pathogens has moderate effects in the cow but may be lethal to a fetus, with the fetal infection inducing lymphoid hyperplasia and a rise in immunoglobulins [46]. Histological alterations in lymphoid organs were observed in both BLV negative and positive fetuses. BLV positive fetuses presented hyperplasia in all thymus samples (n=10), 7 of 8 lymph nodes, and in white pulp of all of 13 spleen samples. BLV negative samples showed hyperplasia in 38 of 53 thymus samples analyzed, 20 of 30 lymph nodes, and in white pulp of 23 of 28 spleen samples.
BLV positive fetuses presented mononuclear inflammatory infiltrate in all liver samples (n=10), 11 of 13 lung samples, 4 of 12 heart samples, 8 of 10 adrenal glands, 4 of 8 kidneys, and 9 of 11 brain samples. BLV negative fetuses presented the same type of mononuclear infiltrate in 19 of 21 liver samples, 39 of 47 lung samples, 34 of 47 heart samples, 14 of 28 adrenal gland samples, 12 of 39 kidney samples, and 28 of 34 brain samples. All BLV positive fetuses presented mononuclear inflammatory infiltrate in at least one organ, and this was also true of 64 of the 67 BLV negative fetuses, suggesting infectious abortion associated with other pathogens. Other researchers observed these lesions at high rates in aborted fetal tissues [1,9,13] and the necropsy of BLV seropositive pregnant cows with lymphosarcoma showed two fetuses with neoplastic lesions of organs and reactive lymph nodes [24].
Tissue lesions suggestive of infection were observed in at least one organ of all tested fetuses. It is possible that both BLV and other pathogens were the source of the mononuclear inflammatory infiltrate in organs, as well as the reactivity of lymphoid organs, in all the fetuses the diferential diagnosis was done for other pathogens: in 3 of 13 positive fetuses there was co-infection of BLV and Klebsiella spp., Trueperella (Arcanobacterium) pyogenes, and Streptococcus spp. each identified in a single fetus. Pathogens were observed in 12 of 67 BLV negative fetuses: Neospora caninum in 4, BVD virus in 3, Escherichia coli in 2, and single instances of Trueperella (Arcanobacterium) pyogenes, Brucella abortus, and Staphylococcus aureus. No Leptospira spp. or BoHV-1 was detected (Table 4). In 55 of the fetuses it was not possible to detect any pathogen, a rate similar to that reported in Brazil [10-12], while another study [9] detected specific abortion pathogens in 46.7% of 490 aborted bovine foetuses.
Table 4: Other pathogens identified in BLV positive and negative fetuses.
| Pathogens | Positive BLV | Negative BLV |
|---|---|---|
| Cases (%) | Cases (%) | |
| Trueperella (Arcanobacterium) pyogenes | 1 (7.69) | 1 (1.49) |
| Brucella abortus | 0 | 1 (1.49) |
| Klebsiella spp. | 1 (7.69) | 0 |
| Streptococcus spp. | 1 (7.69) | 0 |
| Neospora caninum | 0 | 4 (5.97) |
| Bovine Diarrhea Virus | 0 | 3 (4.48) |
| Escherichia coli | 0 | 2 (2.98) |
| Staphylococcus aureus | 0 | 1 (1.49) |
| Bovine Herpesvírus -1 | 0 | 0 |
| Leptospira spp. | 0 | 0 |
| Subtotal/Total (%) | 3/13 (23.07) | 12/67 (17.91) |
Considering that BLV causes leukemia and lymphosarcoma in young and adult animals, and that this virus can cause immunosuppression and vulnerability to pathogens that can cause abortion, the BLV transplacental transmission rates in Brazilian herds show the necessity for preventive programs to avoid fetal infection. Further studies with a bigger sampling are needed to understand whether BLV is a causative or predisposing agent of bovine abortion.
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Thanks to Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, São Paulo, Brazil) for Research Grant processes 2009/13864-8 and 2012/01033-7. To FAPESP for TT-3 Scholarship (Capacitação técnica nível III): 2012/06276-5. To CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior Brasil – código de financiamento 001), for Masters Grant.A.
Citation: Montanari KCS, Fusuma MM, Lacerda AMD, Okuda LH, Pituco EM, de Carvalho AF, et al. (2019) Chronic Lymphocytic Leukemia in a Black African Man: A Cameroonian Case Report. J Leuk 7:253. doi: 10.35248/2329-6917.7.253
Received: 31-Dec-2018 Accepted: 21-Jan-2019 Published: 30-Jan-2019 , DOI: 10.35248/2329-6917.19.7.253
Copyright: © 2019 Montanari KCS, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.