Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research - (2021)Volume 14, Issue 10
The GenBank database yielded a total of fifteen (15) Growth Arrest and DNA-Damage-Inducible protein 45 Alpha (GADD45A) gene nucleotide sequences from goats (5), sheep (5), and cattle (5). The non-synonymous single nucleotide polymorphism of the GADD45A gene of amino acid substitution for cattle, goats, and sheep was checked using the PROVEAN tool. Particular amino acid replacements were found to be neutral/beneficial, while others were shown to be deleterious/harmful. Tajima’s test and phylogenetic association for cattle, goats, and sheep were performed using Molecular Evolutionary Genetics Analysis (MEGA) 5.0. The Tajima’s test (D) yielded good results for all of the species, indicating purifying selection. Because livestock improvement is dependent on the knowledge of genotype and phenotype interaction, the phylogenetic connection revealed a high level of intermingling across and within species, therefore selecting this gene could be valuable in establishing a breeding program for livestock development.
Gene; Substitution; Polymorphism; Non-synonymous; Phylogenetic
GADD45A is a potential contender gene for meat quality traits that controls Quantitative Trait Loci (QTL) [1]. GADD45A is an important stress sensor that regulates the cellular response to a variety of stressors, as well as genotoxic and oncogenic pressures [2]. GADD45A protein has recently been discovered to have a substantial part cutting-edge inheritable factor precise energetic genetic material demethylation through adipose resulting to mesenchymal stem cell development [3]. For a variety of species, recent breakthroughs in great output knowhow have created huge volumes of genome amino acids arrangement and genotyping facts. Experimental techniques make identifying functional SNPs from a pool comprising both functional and neutral SNPs difficult. As a result, computational forecasts have developed critical in assessing the illness connected influence of non- synonymous single-nucleotide variations found through exome sequencing [4]. A numeral of computational procedures have remained established to anticipate the practical outcome of a non- synonymous Single-Nucleotide Polymorphism (nsSNP), which is a single-nucleotide alteration in a gene’s protein coding section that results in an Amino Acid Substitution (AAS) [5]. The purpose of this study was to determine the non-synonymous, selection test, and phylogenetic relationship of the GADD45A gene in cattle, sheep, and goats.
A total of fifteen (15) GADD45A gene nucleotide sequences were obtained from the GenBank (NCBI) database, including goat (5), sheep (5), and cattle (5). (www.ncbi.nlm.nih.gov). The sequences’ Genbank accord numbers were XP 005678524, XP 017906267, XP 017908001, XP 005684830, and XP 005682834 (goat), XP 012043817, XP 012027084, XP 004005614, XP 012008553, and XP 012007133 (sheep), NP (cattle). The GADD45A gene of diverse species was aligned, translated, and compared using Clustal W, as designated by Larkin et al. with IUB replacement medium, breach exposed penalty of 15, and breach leeway penalty of 6.66. Missense mutations remained discovered in silico via Protein Variant Effect Analyzer (PROVEAN) by means of a verge value of -2.5. PROVEAN uses BLASTP (ver.2.2.25) with an E-value cutoff of 0.1 to collect a established of homologous and vaguely connected arrangements of amino acids of genetic materials from the gene bank. The CD-HIT tool (ver.4.5.5) was used to cluster the sequences based on an 80 percent sequence identity to reduce redundancy [6]. The variation is anticipated to be harmful if the PROVEAN score is fewer than or equivalent to a -2.5 threshold) [7]. By means of the Maximum Likelihood technique grounded on the Equivalent Contribution classical, the evolutionary history was inferred [8]. This is the tree with the highest log likelihood (-708.1487). Following the twigs is the proportion of trees in which the connected taxa bunched composed. The preliminary tree(s) for the empirical hunt remained by design created through put on the Neighbor-Join and BioNJ procedures to a medium of pairwise distances forecast via the JTT model, and then picking the topology per the uppermost record prospect value. A total of 13 amino acid arrangements were studied. Breaches and misplaced facts were detached from entire locations. The final dataset contained a total of 27 locations. MEGA7 was used to conduct evolutionary analysis [9]. The physical and chemical changes remained calculated by means of the Poisson correction method [10], and are measured in amino acid substitutions per site. A total of 13 amino acid sequences were examined. Breaches and misplaced facts were removed from all positions. The final dataset contained a total of 63 locations. MEGA7 remained used to carried out physical and chemical changes investigation [11]. MEGA7 was used to calculate the Tajima test statistic [12]. Breaches and misplaced facts were detached from entire opinions in the dataset (Whole removal choice). The acronyms used remain as trails: m=number of sites, S=Number of segregating sites, ps=S/m, Θ=ps/a1, and π=nucleotide diversity. D is the Tajima test statistics [12].
Tables 1-3 show the functional analysis of coding nsSNPs in the Gadd45 gene in sheep, goats, and cattle, respectively. The functional study of the Gadd45 gene of Sheep variations (G11L, K15A, T23K, L36K, F43D, E49P, and G56V) revealed that the coding nsSNP (G11L, K15A, T23K, L36K, F43D, E49P, and G56V) appeared to be detrimental, while the rest of the variant appeared to be helpful. The functional investigation of coding nsSNPs in the GADD45 gene of goat variations (E14Q, G20S, N48K, V54P, and C57V) revealed that some were harmful, while others were advantageous. The functional investigation of coding nsSNPs in the Gadd45 gene of cattle variations (Q31L, A44Y, N48K, D52V, V54P, C57V, and A60C) revealed that some of them were harmful, while others were advantageous. The advantageous variant gives reason for optimism for future selection. Table 4 shows Tajima’s neutrality test results. The Tajima’s test (D) provided positive results aimed at entirely three classes of the animals, as well as a significant level of nucleotide diversity. Figure 1 depicts the phylogenetic relationship. The level of intermingles among the three species was revealed.
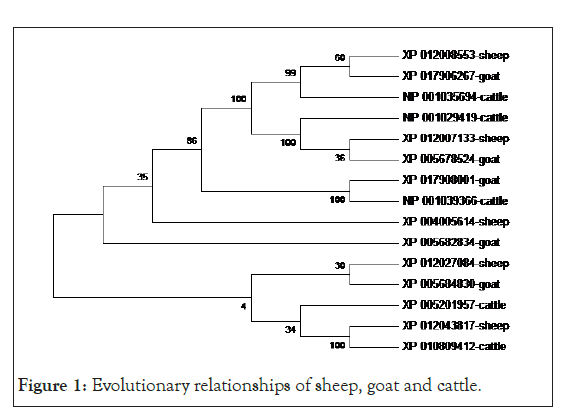
Figure 1: Evolutionary relationships of sheep, goat and cattle.
| Species | M | S | Ps | Ѳ | Π | D |
|---|---|---|---|---|---|---|
| Sheep | 5 | 159 | 0.993750 | 0.477000 | 0.890000 | 6.597472 |
| Cattle | 5 | 135 | 1.00000 | 0.480000 | 0.881481 | 6.368302 |
| Goat | 5 | 131 | 0.992424 | 0.476364 | 0.86182 | 6.261456 |
Note: m=number of sites, S=Number of segregating sites, ps=S/m, Θ=ps/ a1, and π=nucleotide diversity. D is the Tajima test statistic.
Table 4: Results from Tajima’s neutrality test.
GADD45A is a fissionable protein tangled in genomic steadiness, genetic material reparation, and cell development retardation. It also plays a role in active DNA demethylation [13]. Tables 1-3 illustrate the functional study of non-synonymous single nucleotide polymorphisms in sheep, goats, and cattle, which revealed both beneficial/neutral and deleterious/harmful effects. Protein structure, function, interaction, and chemical properties may be altered by beneficial/neutral amino acid changes.
Protein destabilization, enzyme function, and disease susceptibility may all be affected by deleterious/harmful amino acid changes. The beneficial/neutral amino acid alteration could give future breeding programs hope [14]. Because the primary goal of an animal breeder is to select the greatest animals to be the parents of the following generation. As a result, non-synonymous amino acid substitution (nsSNP) of deleterious/harmful variations should be considered while improving the GADD45A gene since they may increase the frequency of disease allele in the ovine, caprine, and bovine populations. As a result, a concerted effort should be made to boost the GADD45A gene, with the goal of transforming the ovine, caprine, and bovine populations. Table 4 shows that Tajima’s trial for assortment yielded helpful results aimed at all of the classes of animals studied. This remains a marker of balance selection, which means a population maintains numerous alleles at a locus. This could help with selection against potentially harmful amino acid substitution variations. Figure 1 depicts the phylogenetic relationship. The gene has been discovered to be interrelated in all three species, which could be attributable to the proximity of a inheritable factor amid animals with four compartment stomach by way of an outcome of a verge in physical and chemical variations owing to the similar assortment forces that animals with four compartment stomach face through physical and chemical variations [15]. The author also speculates that the gene’s close proximity or interaction may be attributable to evolutionary changes in the morphological, physiological, and developmental characteristics of interest.
GADD45A is a fissionable protein that plays a vital function in active DNA and is implicated in genomic stability, DNA repair, and cell growth regulation. Because livestock improvement is dependent on knowledge of genotype and phenotype interaction, selecting this gene could be valuable in establishing a breeding program for livestock development.
Citation: Dauda A, Okon B, Ibom LA (2021) Bio-Computational Analysis of Growth Arrest and DNA-Damage-Inducible Protein 45 Alpha Gene of Cattle, Sheep and Goat. J Proteomics Bioinform.14:556.
Received: 07-Oct-2021 Accepted: 21-Oct-2021 Published: 28-Oct-2021 , DOI: 10.35248/0974-276X.21.14.556
Copyright: © 2021 Dauda A et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.