Journal of Antivirals & Antiretrovirals
Open Access
ISSN: 1948-5964
ISSN: 1948-5964
Research Article - (2021)
We previously predicted Nsp2 Corona virus protein as RNA topoisomerase through amino acid homology among Vibrio haemolytica DNA topoisomerase IA/IV as well as DNA primase, DNA gyrase and bi-subunit Trypanosoma brucei DNA topoisomerase IB. Many DNA topoisomerase I/III have RNA topoisomerase activity and such ubiquitous enzymes are conserved and involved in the regulation of replication and transcription. We have checked here mutational profile of Nsp2 RNA topoisomerase analyzing >10000 orf 1a 4405 amino acid length Corona virus polyprotein. Mutant proteins were selected by BLAST search having 99.84% sequence similarity and 181- 818 aa portion NsP2 protein (protein id. QIU82057) was analyzed using CLUSTAL Omega software. We found 26 different mutations where most changes were selected at Isoleucine and Alanine into Valine or Leucine into Phenylanaline pinpointing conserved nature of the Corona virus RNA topoisomerase. Major nonsense very abundant mutations were found at I120F (Isoleucine to Phenylalanine). Other important mutations were R27C, I198V, T85I, L410F, I559V and P583S. The I120F mutation was abundant in Australian isolates and its spread was seen in the Bangladesh and other countries like USA. We suggest that abundant I120F mutation of Nsp2 Topoisomerase may increase transmission of Corona virus by stabilizing RNA structure for efficient virus packaging. Interestingly, such mutations were found in association of D614G mutation of Spike protein, known to >70% increase infectivity. On the contrary, all P583S Nsp2 mutants analyzed had no concurrence D614G spike protein mutation. Many silent mutations (5-7) were detected by genome wide analysis but no N501Y Spike protein mutation. This is first report that predicts a link of greater Corona virus transmission with Nsp2 protein I120F mutation and may be important to discover new anti-viral drugs.
Corona virus; Nsp2 protein; RNA topoisomerase; Point mutation; Multi-alignment; Higher transmission; Australian isolates
The COVID-19 pandemic has caused more than 1.8 million deaths and over 50 million confirmed cases worldwide, according to the World Health Organization. Corona viruses are large single-stranded positive sense RNA viruses. There are 26 putative coding regions which cover about 98% of the 29.9 kb SARS-CoV- 2 genome [1]. Approximately two-thirds of the genome at the 5' side encodes the two large non-structural polyproteins (orf1ab, 7096 aa and orf1, 4405 aa) in same reading frame and one-third at the 3' side encodes four structural proteins: spike glycoprotein (S), envelope (E), membrane (M), and nucleocapsid (N) [2,3]. Polyproteins are degraded into sixteen proteins like proteases (Nsp3 and Nsp5), RNA-dependent RNA polymerase (Nsp12), rRNA methyl transferase (Nsp13, Nsp16 etc.) and RNA topoisomerase (Nsp2) [4,5]. We have suggested that peptide vaccine from RNA topoisomerase and rRNA methyl transferase may be useful as many mutations are accumulating in the mostly targeted Spike protein of Corona virus [6-8].
COVID-19 is related to SARS and MERS Corona viruses [9]. The mutation rate in the SARS Corona virus genome was estimated to be 0.80-2.38 × 103 nucleotide substitution per site per year which is in the same order of magnitude as other RNA viruses [10,11]. In other related infectious bronchitis virus, it was estimated to be 0.3-0.6 × 10-2 per site per year [12]. Recent estimates suggest that circulating SARS-CoV-2 lineages accumulate nucleotide mutations at a rate of about 1-2 mutations per month [13]. The new variant strain, VUI-202012/01, may be up to 70% more transmissible and could increase the R number by 0.4. Mutations are common to the RNA virus and usually occur due to combination of natural drift and pressure from the human immune system [14].
Proteins are folded into highly complex 3-D shapes, depending on the interactions between different amino acids side chain in the same string [15]. Changing an amino acid of positively charged with one for a negatively charged one, will change the shape and active site. DNA topoisomerases are 2 types called type I and type II DNA topoisomerases which change linking number by 1 or 2 respectively [16]. Such enzymes have nuclease activity, swivel strand passing activity and ligation activity [17,18]. Some type I topoisomerases have shown with RNA topoisomerase activity [19]. We predicted a RNA topoisomerase activity in the Corona virus genome to resolve the knots of dsRNA to synthesis mRNAs and virus packaging of (+) sense ss-RNA [20]. We have shown unknown Nsp2 protein of Corona virus polyproteins has sequence similarities with Vibrio haemolytica DNA topoisomerase I/IV [4]. We also able to dissect the 5’-end ATPase, middle topoisomerase and 3’-end RNA-binding domains by similarity search with amino acid sequences of bisubunit Escherichia coli DNA Gyrase and bi-subunit Trypanoma brucei DNA Topoisomerase IB where ATPase, DNA-binding and topoisomerase activities located in different subunits and domains [4,18]. Thus, we want to check the mutation motifs of Np2 protein from different countries and their abundance in the GenBank database to predict virus stability and infectivity. Such study with Corona virus spike protein gave monumental result where D614G mutation of spike protein showed abundant and had increase in infectivity due to its better interaction with ACE-2 receptor of human cells [21-23]. We found at least 26 different point mutations in the Nsp2 RNA topoisomerase and I120F mutants were abundant in the database. We also found most mutations of Nsp2 proteins associated with D614G spike protein mutation except Nsp2 P583S mutants [24].
Identification of normal and mutant orf1a 4405 amino acid polyprotein amino acid sequence of Corona virus (COVID-19)
We selected Accession number QIU82057 China isolate as normal strain thinking such isolates originate in China first (Isolation date 22-01-2020). When we BLAST searched 648 aa Nsp2 sequence, >4000 sequences are 4405 amino acids with 100% identical and few hundred are 99.84% suggesting Nsp2 mutants [25]. To select more mutants from NCBI GenBank database, we write orf1a, corona virus and country name in the portal www.ncbi.nlm.nih.gov/protein and selected few 4405 aa length orf1a mutant polyprotein sequences. Those mutants were not abundant (1-8) and would take long time during BLAST. So we reduced the search algorithm parameter (at the 2/3 position of the NCBI portal) into 10. If 100% were mutants using such parameter as 10, then we increased algorithm into 100 to 250 to 5000 depending on 100% similarity outcome to trap all mutants [5].
Multi-alignment and selection of mutants and their spread in the different countries
We selected 638 amino acids length Nsp2 sequences and multialign was performed by CLUSTAL-Omega software and from the mismatch of amino acid sequence we got mutant position [26,27]. We then selected phylogenetic relation and phylogenetic picture gave the cluster of normal verses mutants. Such study also showed the different countries in the same mutant cluster indicating the spread from one country into another.
Detection of specific mutant population and spread into different countries
To check the mutant population in the GenBank database, we put different mutant Nsp2 amino acid sequence into NCBI BLAST for protein (www.ncbi.nlm.nih.gov/blast) and 100% similarity sequences are mutants and 99.84% are normal. We selected and opened all mutants amino acid sequences and checked their country origin of individual Accession number and date of collection collected. Most mutants protein id were given in the analysis so that anyone can check any ofr1a sequence for further analysis. Further, RNA sequencing error might be happen and usually such sequence should be removed from database. Thus, those mutants appeared in many countries with high density and were sequenced by different reputed laboratories could be assigned as authentic.
Genome analysis and relation into spike protein D614G hyper-infectivity mutant
We first open orf1a amino acid sequence and then we selected individual accession number to find the spike protein sequence. From the downstream Corona virus sequence, we selected nucleotide sequences of Nsp2, S proteins or full length sequence. Then, we analyzed the sequences by Clustal-omega and Multalin softwares.
Genome analysis to get nucleotide substitution and amino acid changes
We first download the genome sequence from Genbank database and compared with standard Corona virus genomic sequence with Accession no. MT079853. Two sequences were aligned by Multalin software and mismatch point noticed. To get the amino acid position and proper codon change in the alignment, we detected position of mutation and subtracted 225 nt UTR and then divided by 3 to get amino acid position. Then, we checked the corresponding position in the Nsp2 (say) in the ORF1a amino acid sequence and scanned the codon table to assure the amino acid [28]. If amino acid matched then we deducted 180 aa of Nsp1 amino acid to get the Nsp2 amino acid position. The position of sixteen Nsp non-structural proteins are known from the genome database. In case of structural N, M N, E proteins, corresponding nucleotide position was checked from database individual accession number to assure if it fall in that nucleotide sequence position and deducted nucleotides from the N-protein (say) start position and resulted nucleotide then divided by three to get N-protein amino acid position. Then, we checked the codon table to assign amino acid to fit normal verses mutant amino acid. Then, individual protein sequence (say N protein) was compared normal vs. mutant to assure correct interpretation and position by Seq-2 BLAST search. The normal genome RNA sequence Accession number MT079853 was taken as standard and normal orf1a protein amino acid sequence with protein id. QIU82057 was taken as standard.
We searched for Corona virus Nsp2 RNA topoisomerase mutants by multi-alignment and phylogenetic analyses (www. ncbi.nlm.nih.gov/blast). We searched orf1a, Covid-19 and name of country into NCBI portal (www.ncbi.nlm.nih.gov/protein) to get as many as sequences. Figure 1 showed the sequence of Nsp2 RNA topoisomerase and its position onto Corona virus 29.9 kb ss-RNA genome. The position of Spike protein at the 3’-end of the genome was also identified in Figure 1. Figure 2 showed the region of alignment of different Nsp2 proteins where maximum mutations were observed. The phylogenetic relation of mutants vs. normal Nsp2 proteins was shown in Figure 3 indicating about 1/3 of the Nsp2 proteins are mutants. We used China isolates from December 2019-January 2020 to get standard amino acid sequence of Nsp2 protein (Accession nos. YP_009725295, NC_045512, MT079853). Abundant mutations were obscure as we predicted that Nsp2 RNA topoisomerase was conserved and such enzyme was involved in the regulation of Corona virus replication and transcription. So, we extended our studies to different countries, selecting NCBI GenBank database search using country name, orf1a and covid-19. When such Nsp2 mutants were analyzed, we found I120F Nsp2 mutant were abundant. Such multi-alignment portion was shown in Figure 4 and phylogenetic relation was shown in Figure 5 predicting that I120F mutants were very abundant in the database and thus hyper-infective similar to D614G (Aspartic acid vs. Glycine) mutant of spike (S) protein of Corona virus. It appears Australian Corona virus isolates were mostly I120F Nsp2 mutant and we selected more mutants from database. Analysis showed that Bangladesh Corona virus isolates were clustered with Australian isolates suggesting Corona virus spread between two countries (Figure 6). All the 26 mutations of Nsp2 protein was tabulated in Table 1.
| Serial. No. | Mutation and amino acid position | Population of the nsp2 mutants | Accession number in the Genbank database | Nucleotide change in the Corona virus genome | Protein id ORF1a of Corona virus polyprotein | Origin of the Corona virus | Spike mutation at 614 amino acid |
|---|---|---|---|---|---|---|---|
| 1 | F10=L | 8 | MW420694 | U>C | QQQ78828 | USA | G614 |
| 2 | L24=F | 9 | MW023471 | U>C | QNU11135 | USA | G614 |
| 3 | R27=C | 14 | MW191505 | C>U | QOU99215 | India | D614* |
| 4 | T45=I | 1 | MW166221 | C>U | QOQ72554 | India | G614 |
| 5 | T85=I | >80 | MW309425 | C>U | QPJ77271 | Canada | G614 |
| 6 | I120=F | >4000 | MW183934 | A>U | QOT47645 | Australia | G614 |
| 7 | A159=V | 35 | MW420612 | C>U | QQQ79846 | USA | G614 |
| 8 | V198=I | 12 | MW81828 | G>A | QOS50880 | India | D614* |
| 9 | S248=N | 1 | MN994467 | G>A | QHQ71962 | USA | D614* |
| 10 | A249=V | 2 | MT883503 | C>U | QNC69817 | Serbia | G614 |
| 11 | V259=I | 1 | MW197485 | G>A | QOW17104 | USA | G614 |
| 12 | E264=K | 5 | MT846524 | G>A | QMU92722* | USA | E614 |
| 13 | I281=V | 2 | MT450961 | A>G | QJR85580 | Australia | D614* |
| 14 | I293=V | 1 | MW030272 | A>G | QNV50417 | Peru | G614 |
| 15 | A294=V | 1 | MW077498 | C>U | QOI11882 | USA | G614 |
| 16 | A302=V | 4 | MT929131 | C>U | QNL12829* | USA | G614 |
| 17 | A336=V | 8 | MW309470 | C>U | QPJ77797 | USA | G614 |
| 18 | S366=P | 7 | MW420612 | U>C | QQG77846 | USA | G614 |
| 19 | A375=T | 1 | MW065478 | G>A | QOF21676 | USA | G614 |
| 20 | R380=H | 12# | MW420694 | G>A | QQG78828 | USA | G614 |
| 21 | I393=V | 4 | MW206450 | A>G | QOW84791 | USA | G614 |
| 22 | L410=F | >30 | MT928991 | G>T | QNL11149 | USA | G614 |
| 23 | V425=I | 1 | MW280543 | G>A | QPG00731 | USA | G614 |
| 24 | A476=V | 4 | MW280547 | C>U | QPG00779 | USA | G614 |
| 25 | Q483=R | 3 | MW010238 | A>G | QNT08568 | USA | G614 |
| 26 | V485=I | >30 | MW077467 | G>A | QOI11510 | USA | G614 |
| 27 | I559=V | >64 | MT407655 | A>G | QKE23290 | China | D614* |
| 28 | P583=S | >64 | MT407655 | C>U | QKE23290 | China | D614* |
| 29 | I605=V | 1 | MW115047 | A>G | QOL01936 | USA | G614 |
| 30 | G638=S | 1 | MT093571 | G>A | QIC53203 | Sweden | D614* |
Note: * means normal amino acid at 614 position. # means some partial sequences have mutant allele but not included here.
Table 1: Mutations and spread of Nsp2 RNA Topoisomerase of Corona virus.
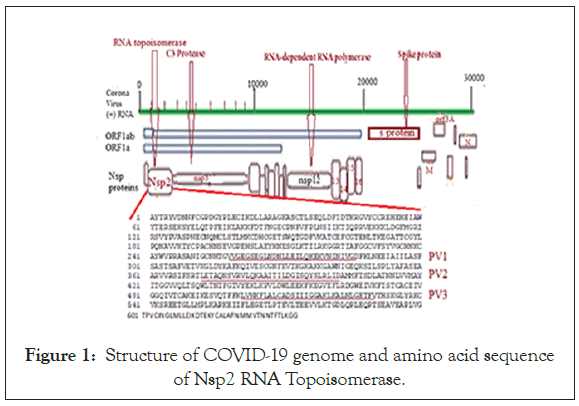
Figure 1: Structure of COVID-19 genome and amino acid sequence of Nsp2 RNA Topoisomerase.

Figure 2: Multi-alignment of Nsp2 protein of different Corona virus isolates showing cluster of mutations between 240-300 amino acids.
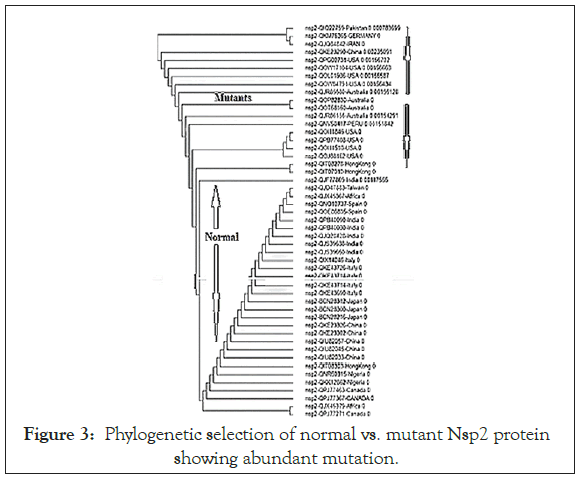
Figure 3: Phylogenetic selection of normal vs. mutant Nsp2 protein showing abundant mutation.

Figure 4: Multi-alignment of Nsp2 protein of Corona virus isolates showing I120F mutation and its high transmission into Australia.
Figure 5: Phylogeny of I120F Corona virus Nsp2 protein mutant in Australian isolates between June-November collection dates.
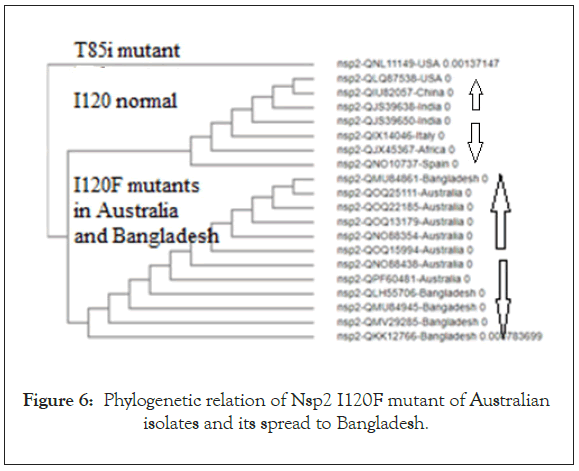
Figure 6: Phylogenetic relation of Nsp2 I120F mutant of Australian isolates and its spread to Bangladesh.
We searched individual mutant Nsp2 amino acid sequence by BLAST to get the population density of individual mutant (Table 1, Column 3). We found that I120F Nsp2 mutant were very abundant (>4000 mutants) with respect to other mutant strains. Then, we checked if spike protein D614G mutation was occurred with Nsp2 I120F mutation. Our first such analysis was shown in Figure 7 indicating abundant D614G mutation in our Nsp2 I120F isolates. Such analysis detected Spike protein S477N mutation in four Australian isolates with Accession nos. MW183334, MW153105, MW156105 and MT971831. We also found T22I Spike mutation in MT582454 Germany isolate (protein id. QKM76366) with no D614 mutation and two V3G and L5F extra mutations in Canada isolate (protein id. QPJ77464) with D614G mutation (data not shown). Further, to confirm our hypothesis of higher transmission of I120F mutants, we searched more mutants from Australia (accession nos. MW154848, MW154818, MT971202, MT969734, MW184478, MW156737, MT971875, MT970805, MT970517, MW153100, MW185536, MW15981, MW156921, MW185678, QW156991, MT972745, MW157103, MW184445, MW185110, MW277558, MW154653, MT971287, MW154664, MW321131, MW184624, MW185253, MW184329). Similarly, we found few United States isolates with I120F mutants (Accession nos. MW075791, MT811489, MW056129, MW276575) and one Bangladesh isolates with I120F mutant. All I120F mutants from Australia, USA and Bangladesh discussed above had D614G mutation in the spike protein of Corona virus. Further we extended our search to prove the Australian I120F Nsp2 mutants transmitted into Bangladesh. For that we used genomic mutant sequence (901 gggttgaaaa gaaaaagctt gatggcttta tgggtagatt tcgatctgtc tatccagttg) for BLASTn and received >4500 I120F mutant sequences referring ORF1ab 7096 aa large polyprotein. Although most sequences were isolated in the Australia, we identified 16 Bangladesh I10F Nsp2 mutants with D614 spike protein mutation together to prove that I120F Nsp2 mutants indeed spread into Bangladesh. The Accession numbers of the Bangladesh double mutant isolates were: MT576639, MT846005, MT731734, MT576641, MT539159, MT664171, MT847211, MT661278, MT856455, MT676414, MT581426, MT667351, MT846011, MT731735, MT578017, and MT761657. We expected to get such mutant isolates of Indian origin, specifically from Kolkata but failed during such search but few United States isolates obtained as discussed above.
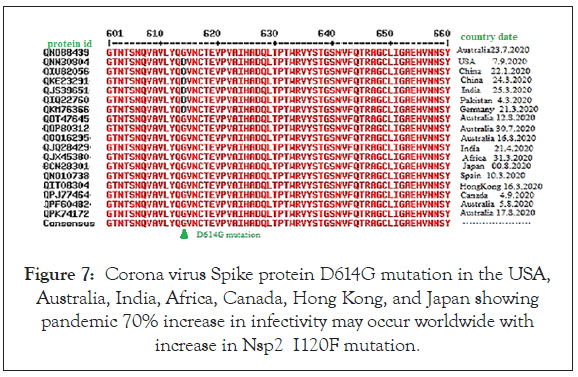
Figure 7: Corona virus Spike protein D614G mutation in the USA, Australia, India, Africa, Canada, Hong Kong, and Japan showing pandemic 70% increase in infectivity may occur worldwide with increase in Nsp2 I120F mutation.
We found mostly single amino acid Nsp2 mutant and thus we wanted to check the relation of other Nsp2 mutants with concurrence D614G S-protein mutation. The data shown in Figure 8, where we found most Nsp2 A336V mutants had D614G mutation as well as in the some A336 normal isolates. Nsp2 T85I mutants from the USA were limited but all appeared to be associated with concurrence D614G spike protein mutation as shown in Figure 9. Thus, we predicted that abundant D614G spike protein mutation might be maximum and other Nsp2 mutations might be occurred lately (June, 2020) but rapid spread of I120F Nsp2 mutants likely increased infectivity together in Australian isolates. Such mutant isolates were abundant in the Australia and Bangladesh as well as might be happening in the most Asian countries! No N501Y mutation in the Spike protein detected and such mutation was detected in the United Kingdom recently with some Corona virus spread. However, Nsp2 P583S mutants had no spike protein mutation like D614G (Figure 10). We also showed that L410F Nsp2 mutants had D614G spike protein mutation (Figure 11) [29]. Such analysis also detected few extra mutations (S704L and M740I; Accession no. MW276622) and (Q321Z; Accession no. MW276643) in the Spike protein (data not shown). We have to wait and see as more genetic data to be deposited in the GenBank specifically from England. We detected two Nsp2 G638S and S248N mutants by checking data of orf8 and orf1ab mutants published by Kailany and co-workers [30]. Because few sequences (Accession nos. LC522972/3/4/5) used in this study were removed from database, such study depend on the authenticity of sequencing [31].
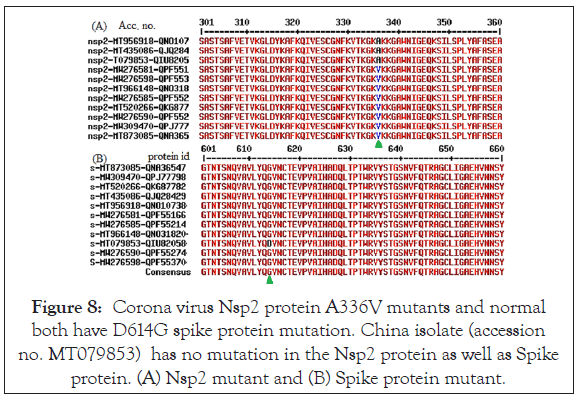
Figure 8: Corona virus Nsp2 protein A336V mutants and normal both have D614G spike protein mutation. China isolate (accession no. MT079853) has no mutation in the Nsp2 protein as well as Spike protein. (A) Nsp2 mutant and (B) Spike protein mutant.
Figure 9: US-isolates of Nsp2 T85I Corona virus mutant have D614G spike protein mutation. Accession no. MT079853 is control China isolate and rests are T85I mutants.

Figure 10: Analysis of covid-19 Spike protein D614G mutation in Nsp2 P583S mutant isolates. Surprisingly, no D614G mutation in the spike protein was detected.
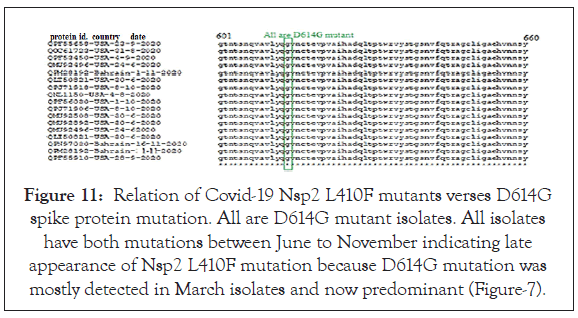
Figure 11: Relation of Covid-19 Nsp2 L410F mutants verses D614G spike protein mutation. All are D614G mutant isolates. All isolates have both mutations between June to November indicating late appearance of Nsp2 L410F mutation because D614G mutation was mostly detected in March isolates and now predominant (Figure-7).
We also got few P812L spike protein mutation in Indian isolates (Figure 12). Such P812L spike protein mutation was found in the other countries like USA (protein ids. QOY58922; QPB77589; QPI70053) and Egypt (protein id. QLQ87540). Many other similar Indian mutants also reported (see, protein ids. QPZ33510, QNN88094, QOQ72556, QNN88166, QNL35902 and QPB18136). We detected T45I Nsp2 new mutation in an Indian isolate (Accession no. MW166221; ORF1ab protein id. QOQ72554) and previously identified T85I and V485I mutations in the US isolates (Accession no. MW252136 and MW292161 respectively).

Figure 12: Detection of two Nsp2 G638S and S248N mutants by checking data of orf8 and orf1ab mutants published by Kailany et al. Because few sequences (Accession nos. LC522972/3/4/5) were removed from database, such study depend on the authenticity of sequencing.
We also detected 8 mutations in the Corona virus genome (Accession no. MW023471; 29811bp) where one L24F missense mutation (TTT=CTT) and one silent mutation at 577 aa Valine of Nsp2 (GTC=GTT), one silent mutation at 106 aa (Phenylalanine) of Nsp3 (TTT=TTC) and at 2201 aa of Nsp3 protease (TTT=TTC), one missense mutation (TCA=TTA) in the Nsp4 protein (S111L) and three adjacent missence mutations in the N-protein AAA=AGG (Arginine203 to Lysine) and GGA=CGA (Glycine204 to Arginine). T will be U in case of RNA genome of Corona virus (Table 1). Never the less, such bioinformatics analysis is worthy important to predict the severity of Corona virus transmission in relation to Nsp2 RNA topoisomerase as well as in the other important Corona virus proteins like Spike protein which binds efficiently to ACE-2 receptor of human cells for virus integration (Figures 13 and 14).

Figure 13: Detection P812L mutation of Spike protein of Corona virus in India with no mutation in the Nsp2 RNA topoisomerase. Accession no. MT940464 Indian isolate was reported three Spike protein mutations (L56F, R346T and Q612H) but no Nsp2 mutation was found.
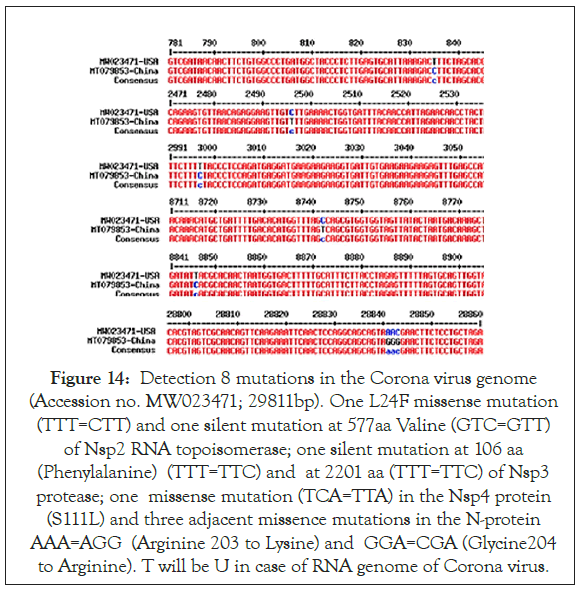
Figure 14: Detection 8 mutations in the Corona virus genome (Accession no. MW023471; 29811bp). One L24F missense mutation (TTT=CTT) and one silent mutation at 577aa Valine (GTC=GTT) of Nsp2 RNA topoisomerase; one silent mutation at 106 aa (Phenylalanine) (TTT=TTC) and at 2201 aa (TTT=TTC) of Nsp3 protease; one missense mutation (TCA=TTA) in the Nsp4 protein (S111L) and three adjacent missence mutations in the N-protein AAA=AGG (Arginine 203 to Lysine) and GGA=CGA (Glycine204 to Arginine). T will be U in case of RNA genome of Corona virus.
We showed 26 mutations and one mutation was very significant with population density and with one or few sequence (s) in the database referred as insignificant. Significant mutations when put on the Nsp2 RNA topoisomerase domains (Figure 15), we found middle RNA topoisomerase domains were unaffected but some mutation in the Carboxy-terminal DNA binding domain would be significant. The R27C mutation at the NH2-terminal might be important as Cysteine helped to make S-S bridge. We presumed I120F mutation might enhanced ATPase activity and enzyme activity through better hydrophobic interaction of phenylalanine pocket but needed crystallographic study for such proof. Never the less rampant isolation of Australian I120F Nsp2 mutant suggested their enhanced transmission in presence of well documented D614G Spike protein mutation (Figure 15).
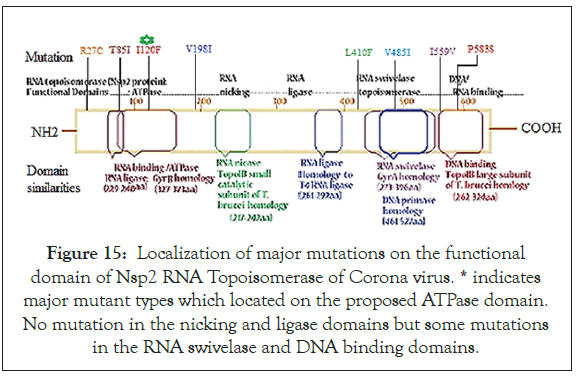
Figure 15: Localization of major mutations on the functional domain of Nsp2 RNA Topoisomerase of Corona virus. * indicates major mutant types which located on the proposed ATPase domain. No mutation in the nicking and ligase domains but some mutations in the RNA swivelase and DNA binding domains.
BLAST search is an important tool to predict function of unknown viral proteins. Corona virus two polyproteins (4405 aa orf1a; 7096 aa orf1ab) are generated in same reading frame and proteolytically have generated sixteen proteins with diverse functions (Nsp1- Nsp16). Nsp12 is RNA dependent RNA polymerase and Nsp3/5 are proteases. Other important functions of such Corona virus non-structural proteins are illuminating lately as rRNA methyltransferases (Nsp13-Nsp16) and RNA topoisomerase (Nsp2) [4,5]. We clearly identified >30 mutations in the Nsp2 RNA topoisomerase which was thought to control Corona virus replication and generation of individual mRNAs required for S, N, M, E structural proteins. However, we found only spread of Corona virus with I120F Nsp2 RNA topoisomerase mutants and such mutation was very much associated with deadly D614G Spike protein mutation. Other important Nsp2 mutants are T85I, V485I, I559V, P583S and L410F with 20-60 individual strains were detected in the GenBank database and likely were important to further study. Such mutation in the RNA virus genome may occur at a rate 1-2 mutation per/month. However, spread of D614G S-protein mutants were overwhelming and it was increased infectivity rate 70%. UK and USA were the worse hit but such mutant had detected in Germany, India and Japan as well as many other countries (Figures 7 and 8). In truth, RNA sequencing of Corona virus genome is costly. So, data from Latin America, Africa and South Asian countries were limited. We have shown that I120F mutation is abundant in the Australia who has deposited many sequences of clinical isolates between July-November, 2020. D614G spike protein mutation was identified in India, Europe, Africa and China. Australian mouse adaptive hyperactive clone (Accession no. MT971831) accompanied with Asn=Ser mutation at position 377 as well as two silent mutations at position 96 (Glu, GAG=GAA) and 613 (Gln, CAG=CAA) in conjunction of D614G mutation of spike protein and I120F mutation of Nsp2 RNA topoisomerase. We have also detected few new Spike protein mutations W152L (accession no. MW217347), A688V (accession no. MW223097) and Q14H (accession no. HT873079) in association with T85I Nsp2 mutation and D614G spike protein mutation (Figure 9). However, Nsp2 P583S mutants had no spike protein mutation like D614G (Figure 10). We also analyzed L410F Nsp2 mutants with D614G spike protein mutation (Figure 11). Such analysis also detected few extra mutations (S704L and M740I; Accession no. MW276622) and (Q321Z; Accession no. MW276643) in the spike protein. Thus, we clearly demonstrated a relation of deadly D614G Spike protein and Nsp2 I120F mutant with Corona virus spread worldwide [23].
Kailany, et al. have reported orf1ab mutations (A1176V, L3606F, E3764D, Y5720C, P6083L) of Corona virus but we found accession numbers (LC522972 and LC522972) were removed from the NCBI database reflecting some error in the sequencing [30]. When Nsp2 protein of rest four sequences were analyzed, we found end amino acid of Nsp2 protein was changed from Glycine to Serine (Figure 12). Kailany, et al. also reported many L84S mutation in the orf8 gene whose function remains elusive (see, accession nos. LC522973, LC522974, LC522975, MT066175, MT188339, MN975262, MN994467).
RNA topoisomerase is an important enzyme of COVID-19 and such research must be augmented [32]. We checked 26 mutants of Nsp2 enzyme but only six mutants with moderate spread would be important and I120F hyper-spread mutant was a concern. High rate mutation accumulation over short time periods have been reported previously in studies of immune-deficient or immune-suppressed patients who are chronically infected with SARS-CoV-2 [27]. Corona virus B.1.1.7 lineage N501Y mutation increased ACE2 receptor affinity to enter into human lung cells [21]. N501Y has been associated with increased infectivity and virulence in a mouse model but data limited in the GenBank. The N501Y mutation cooccurs with several mutations in the N, ORF1a, ORF8 proteins and in S protein 501Y Variant 2, but such ±complete genome data were not deposited to GenBank yet [33,34]. A222V Spike mutation is also immerging in B1.177 lineage and last month its number increased to >7000 in the UK (https://www.cogconsortium. uk/wp-content/uploads/2020/12/Report-1_COG-UK_20- December-2020_SARS-CoV-2-Mutations_final_updated2.pdf). Thus, our data is worthy to predict role I120F mutation of Nsp2 RNA topoisomerase emerging in the Australia [35]. We could not find much I120F mutant in Indian isolates but we should carefully perform RNA sequencing to predict such transmission which may be occurring in the Bangladesh. Recently, many vaccines for Corona virus have developed and vaccination started in Europe and America [36]. We have shown some similarity of Nsp2 RNA topoisomerase to ribosomal L1 protein and with other nonstructural proteins like Nsp13 (L6 and L9) capping mRNA methyl transferase with RNA helicase activity. These suggested such Corona virus proteins may assemble into host ribosome affecting protein synthesis and oxidative phosphorylation [6,37]. Recently, nonotechnology and gene medicine have been given priority to challenge Corona virus spread with 1.7 millions death worldwide [38]. Integrative mutant analyses of SAR-COV-2 genomes from different countries may give clues for host-virus interaction, pathogenesis and development of better therapeutics [14,15,39]. Truly, vaccination research has given priority and vaccination against Corona virus has started [1,36,40,41]. We have proposed three peptide vaccine candidates from Nsp2 RNA topoisomerase [258-285 aa; 372-401 aa and 501-531 aa regions] (Figure 1 underlined sequence). E264K mutation may affect vaccine efficacy in PV1 peptide, similarly A375I mutation may increase vaccine efficacy in PV2 peptide. While no mutation in the PV3 peptide was noticed (NH2-LVNKFLALCADSIIIGGAKLKALNLGETFV-CO 2H) suggesting a good peptide vaccine candidate with many hydrophobic amino acids and glycosylation signals [4]. Corona virus pathogenesis often associated with multidrug-resistant bacterial infection. We have shown that phyto-extracts from Cassia fistula and Suregada multiflora bark and root efficiently inhibited different MDR-bacteria isolated from Ganga River, Chicken and Milk as well as human subject [42-44]. We demand action of different phyto-chemicals should be tested on the replication and transcription of Corona virus.
Recently COVID-19 mutation database (https://www.ncbi.nlm. nih.gov/labs/virus/vssi/#/scov2_snp) has listed more mutations but such mutations have no role in corona virus spread. The new Nsp2 mutations reported are: P17S, E19D, T44S, G47S, E53D, Y61F, P91S, R107S, V126F, P129L, G265S, D268N, L274F, A360T, V364L, R370H, P371I, T436I, K454E, L462F, V485I, E493K, Q496H, K500N, K534R, G548C, P568L, T569R, P585S, P597S, K618N, P624S and T634I etc.
We have detected 30 RNA topoisomease mutants of corona virus and we think I120F mutants have role in higher transmission. Interestingly D614G spike protein and P4715L RdRP mutations have abundant association whereas nsp4 S2874L mutation has some association in corona virus spread. Such epidemiological analysis is important to choose peptide vaccine candidate and may help for new drug design.
We thank our principal Prof. Bidyut Bandhopadhyay for his interest in Corona virus research.
The Author has no conflict of interest.
Citation: Chakraborty AK (2021) Abundant Transmission of Corona Virus Nsp2 RNA Topoisomerse I120F Mutants with Concurrence D614G Spike Protein Mutation in Australia. J Antivir Antiretrovir. S16:001.
Received: 07-Jan-2021 Accepted: 21-Jan-2021 Published: 28-Jan-2021 , DOI: 10.35248/1948-5964.13.s16.001
Copyright: © 2021 Chakraborty AK. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.