Research Article - (2022)
A Method for the Definition of Immunological Non-Response to Antiretroviral Therapy Based on Review Analysis and Supervised
Classification Model
Yong Shuai1,2*,
Hemeng Peng1,4,
Xiaodong Wang1,3 and
Xiaoqing Peng1,3
*Correspondence:
Yong Shuai, Chongqing CEPREI Industrial Technology Research Institute Co., Ltd, Chongqing, 401332,
China,
Tel: +086 156 8362 2221,
Email:
Author info »
Abstract
Background: Immunological Non-Response (INR) accelerated the progression of AIDS disease and brought serious difficulties to the treatment of HIV-1 infected people. The current definition of INR lacked a credible consensus, which affected the diagnosis, treatment and scientific research of INR.
Methods: We systematically analyzed the open source INR related references, used visualization techniques and machine learning classification models to propose the features, models and criteria that define INR.
Results: We summarized some consensus on the definition of INR. Among the features that defined INR, CD4+ T-cell absolute number and ART time were the best feature to define INR. The supervised learning classification model had high accuracy in defining INR, and the Support Vector Machine (SVM) had the highest accuracy in the commonly used supervised classification learning model. Based on supervised learning model and visualization technology, we proposed some criteria that could help to reach a consensus on INR definition.
Conclusion: This study provided consensus, features, model and criteria for defining INR.
Keywords
Immunological non-response; Definition; Review analysis; Visualization; Supervised learning
classification model
Introduction
After Human Immunodeficiency Virus type 1 (HIV-1) entered the human body, it would cause the reduction of CD4+ T lymphocytes (abbreviated as CD4+ T-cell), the gradual exhaustion of CD4+ T-cell,and the destruction of the physical immune function. After effective
combined Antiretroviral Therapy (cART), most People Living with
HIV (PLWH) will be able to achieve virological suppression, and
the CD4+ T-cell count will increase significantly, and the body's
immunity function will gradually recover.
However, there was still about 9%-45% of People Living with
HIV (PLWH) whose CD4+ T-cell count level had not recovered
although they had reached the standard of virological suppression,
and immunological non-response has occurred. These PLWH were
calling PLWH with poor immune reconstitution or Immunological
Non-Responders (INRs) [1]. Corresponding to them were Immunological Responders (IRs), these patients achieved both
virological suppression and CD4+ T-cell count return to normal
value. Since INR would increase the morbidity and mortality of
AIDS-Defining diseases (AD) and Non-AIDS-Defined diseases
(NAD), research on INR had become the focus of current HIVrelated
research, and the relevant contents included the definition,
mechanism and treatment plans of INR [2-6].
Because researchers had different understandings to INR, there was
no consensus on the definition of INR. The features that defined
INR included the CD4+ T-cell count absolute number, CD4+ T-cell
count increase, CD4+ T-cell count growth rate, CD4/CD8 ratio,
time to receive effective cART, Virologic Suppression (VS) time.
Each standard had a significant interval. For example, the CD4+ T-cell count absolute number included 200, 250, 300, 500, and
ART time included 6 months, 1 year, 2 years, 5 years, 10 years, etc.
At the same time, some references still have undefined intervals for the definition of INR. For example, the reference [7] defined INR
with the standard of cd4 count absolute number <200 and IR with
the standard of CD4+ T-cell count absolute number >500, but there
was no definition for the patients with the CD4+ T-cell absolute
number in the interval of (200,500).
The lack of a consensus standard for the definition of INR will
adversely affect the advancement of scientific research and clinical
diagnosis. In terms of scientific research, due to the different
definitions of INR, it was difficult to understand and compare
similar research results, which affected the credibility and reliability
of these research results. In terms of clinical diagnosis, most doctors
would judge whether a patient was INRs based on guidelines [8]
and understanding of INR by using similar indicators or adjacent
time. For example, when a patient came to see a doctor on the
13th month and 12 days after receiving ART, the CD4+ T-cell count
absolute number and the conversion rate at the 12th month in the
guideline were generally used to determine whether the patient was
INRs. This clinical judgment method for non-standard time was not
accurate enough, and it may lead to misdiagnosis or overtreatment.
In order to solve the above problems, through systematic analysis of
INR related references and visualization techniques, we found the
best features to define INR firstly. Then we trained the supervised
learning classification models and obtained the optimal supervised
learning classification model. Finally we proposed some INR
definition criteria for references based on the best supervised
learning classification model, so as to assist doctors and researchers
to carry out diagnosis, treatment and research work.
Methodology
Study design
Data source and definition:
References data: References data was obtained through the websites
of https://pubmed.ncbi.nlm.nih.gov/ and https://www.cnki.net/.
The searching keywords were from Table 1 of the references [2].
The search language of https://pubmed.ncbi.nlm.nih.gov was
English, and the search languages of https://www.cnki.net/ were
Chinese and English.
| |
Number (%) of patients |
|
| INRs |
IRs |
Total |
p |
| (n=459) |
(n=192) |
(N=651) |
| Age (years) |
0.808 |
| Mean |
50.27 |
50.57 |
50.36 |
|
| Median (range) |
51.0(39-60.6) |
51(37-64) |
51(38-62) |
|
| Sex, n (%) |
<0.001 |
| Male |
368(80.17%) |
159(82.81%) |
527(80.95%) |
|
| Female |
91(19.83%) |
33(17.19%) |
124(19.05%) |
|
| Last CD4+T cell absolute number(cells/uL) |
0.435 |
| Mean |
275.61 |
566.17 |
361.31 |
|
| Median (range) |
273(189.5-362) |
564(427-660.5) |
329(229-462.5) |
|
| ART time(months) |
<0.001 |
| Mean |
48.61 |
38.48 |
45.62 |
|
| Median (range) |
46.9(24.85-71.25) |
29.35(16.3-61) |
41.9(21.45-69.55) |
|
Table 1: Basic information of the data set.
Training data of the classification model: Among all the INR
definition related references [1-3,6,7,9-136] retrieved in this paper,
only the reference [59] provided the original INR open sourced
data. The reasons why the original data sources of other references
cannot be obtained included: The data sources website cannot be
opened, the corresponding author needed to be contacted, or only
the data after statistical analysis was provided. In order to ensure
the credibility of the supervised learning classification modeling
results, we used some related data from the electronic medical
record database of Chongqing Public Health Medical Center for
analysis.
Definition of INR and IR in the data source: For open source data
of reference [59], we used the original data. For the data in the
electronic medical record database, we assumed that INRs were
defined as patients who have adopted INR interventions methods
(including the use of Thymalfasin, Thymopentin, Recombinant
Human Growth Hormone, Aikeqing Capsule, Peiyuan Capsule,
Tang Herb Tablets, Mushroom Polysaccharides, etc.) [5] or were
recorded as INR in the cases, and other PLWH were defined as
IRs. Both the INRs and IRs reached the standard of virological
suppression. We combined the open source data and the data from
electronic medical record database into one data set. The basic
information of the data set was shown in Table 1.
First of all, through references analysis, we found the associated
features of the INR definition, and visualized the relationship
between these features and INR. Secondly we proposed hypotheses,
used the supervised learning model to classify INR and IR, and
used cross-validation and grid search in the supervised learning
modeling process to prevent overfitting and obtain the best INR
evaluation model and its corresponding parameters. Finally, we
proposed some criteria on the definition of INR based on the best
supervised learning model. The flowchart of this paper was shown
in Figure 1.
Figure 1: Flowchart of this paper.
In order to facilitate other researchers to rebuild the models in this
paper and carry out more in-depth researches, we open source the
entire modeling process and source code in the paper. The source
code was available in the supplement.
Results
Features Analysis of related to the definition of INR
The references used in the papers were all derived from the
published literature related to INR. We used the method of
literature [2] to systematically analyze the references and sorted out
the definition standards of each paper for INR. Through literature
analysis, we summarized the following consensus regarding the
definition of INR:
1. The HIV-1 antibody test of the patient was positive
2. The patient has received ART for more than 6 months
3. The patient has achieved virologic suppression or reached the
virologic suppression standard in the patient's area
4. The CD4+ T-cell absolute number of the patient failed to return
to normal level
At the same time, we sorted out the features related to the definition
of INR, and its visualization map was shown in Figure 2. It can
be seen from this figure that the features related to the definition
of INR included CD4+ T-cell count absolute number, CD4+ T-cell
count change number, CD4+ T-cell count growth rate, CD4/CD8
ratio, ART time and Virologic Suppression(VS) time.
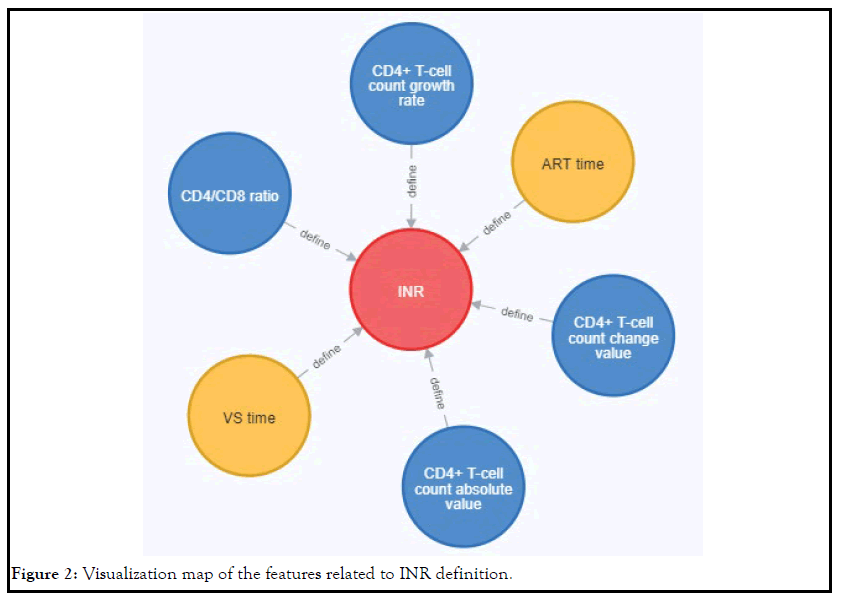
Figure 2: Visualization map of the features related to INR definition.
Features selection
We summarized the six features found in the previous section into
two categories, which were called the medical test features(including
CD4+ T-cell count absolute number, CD4+ T-cell count change
number, CD4+ T-cell count growth rate and CD4/CD8 ratio) and
the time features(including ART time and VS time). The usage
frequency of each feature to define INR was shown in Table 2.
| Feature |
Number of references |
Number(n) |
Percentage(100%) |
| 133 |
100 |
| Medical test feature |
CD4+ T-cell count absolute number |
115 |
86.47 |
| CD4+ T-cell count change number |
25 |
18.8 |
| CD4+ T-cell count growth rate |
13 |
9.77 |
| CD4/CD8 ratio |
4 |
3.01 |
| Time feature |
ART time |
102 |
76.69 |
| VS time |
51 |
38.35 |
Note: There were some combined indicators for the medical test features and time features in some references, such as reference[125] defined INR by the medical test values standard of the CD4+ T-cell count absolute number<200 and the CD4+ T-cell count change number <100, while reference [109] defined INR by the time values standard of ART time> 12 and VS time > 6. We separately counted the combination indicators.
Table 2: INR definition related features and the usage frequency of these features.
It can be seen from Table 1 that the CD4+ T-cell count absolute
number in the medical test features was used the most times, and
the ART time in the time features was used the most times.
By comparing all the medical test features, we can find that the
CD4+ T-cell count absolute number can be obtained every time
when a patient went to the hospital. This feature did not need
to compare with the previous test values (including the absolute
value of the baseline CD4+ T-cell count), but the CD4+ T-cell count
change number and the CD4+ T-cell count growth rate needed
comparison values. Compared with CD4/CD8, the CD4+ T-cell
count absolute number used to define INR had been recognized by
more scholars. Therefore, the CD4+ T-cell count absolute number
was the optimal feature in medical test features for defining INR.
By comparing the time features, we found that the ART time can
be calculated from the time when the patient received ART, which
was easy to calculate, and the calculation standard was uniform.
The acquisition of the VS time required the detection of the viral
load of HIV RNA. However, the current standards for virologic
suppression were not uniform, as shown in Table 3. The acquisition
of VS time required the patient to go to the hospital to check again
after receiving ART. When the patient achieves VS standard after
receiving ART treatment without checking, this time cannot be
accurately obtained. Therefore, ART time was the optimal feature
in medical time features for defining INR.
| No |
HIV-1 RNA viral load (copies/ml) |
References number |
Sum |
| 1 |
20 |
13,20,50,88 |
4 |
| 2 |
40 |
37, 83,93,106 |
4 |
| 3 |
40 to 75 |
102 |
1 |
| 4 |
48 |
100 |
1 |
| 5 |
50 |
7,15,19,21,22,31,34,36,39,43-45,48,49,52-55,61,69,74,77,92,95,97,98,103,105,108,130,134,136 |
32 |
| 6 |
75 |
29 |
1 |
| 7 |
200 |
94 |
1 |
| 8 |
400 |
14,63,135 |
3 |
| 9 |
500 |
136 |
1 |
| 10 |
1000 |
24,60 |
2 |
Table 3: VS Standards of HIV-1 RNA Viral load.
Based on the above analysis, we chosen both the CD4+ T-cell
count absolute number and ART time as the features to define
INR. By selecting the references that only used the CD4+ T-cell
count absolute number and the ART time, we hoped to discover
the relationship among the definitions of INR and Immunological
Response (IR) through visualization techniques. The definition
and relationship of INR and IR were shown in Table 4 and Figure 3.
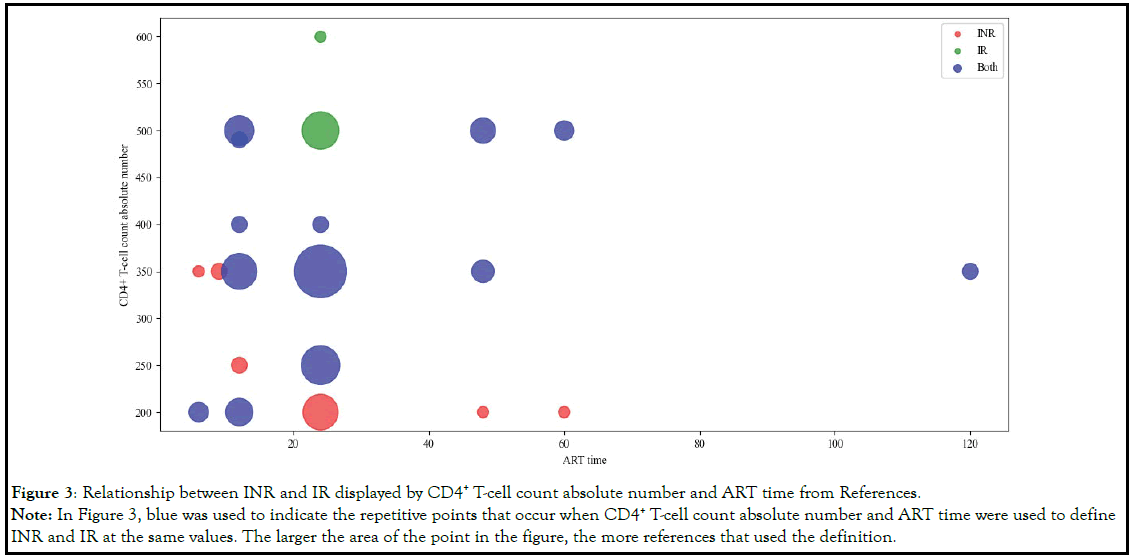
Figure 3: Relationship between INR and IR displayed by CD4+ T-cell count absolute number and ART time from References.
Note: In Figure 3, blue was used to indicate the repetitive points that occur when CD4+ T-cell count absolute number and ART time were used to define
INR and IR at the same values. The larger the area of the point in the figure, the more references that used the definition.
| No |
Definition of INR |
Definition of IR |
Number of overlaps |
| CD4+ T-cell count absolute number |
ART time |
Sum |
References number |
CD4+ T-cell count absolute number |
ART time |
Sum |
References number |
| 1 |
200 |
6 |
2 |
16,61 |
200 |
6 |
1 |
61 |
3 |
| 2 |
200 |
12 |
4 |
6,17,64,135 |
250 |
12 |
2 |
14,64 |
6 |
| 3 |
200 |
24 |
10 |
7,13,15,18-21,68,70,88 |
|
|
|
|
|
| 4 |
200 |
48 |
1 |
11 |
|
|
|
|
|
| 5 |
200 |
60 |
1 |
67 |
|
|
|
|
|
| 6 |
250 |
12 |
2 |
14,22 |
|
|
|
|
|
| 7 |
250 |
24 |
7 |
23-27,35,86 |
250 |
24 |
5 |
24,25,26,27,35 |
12 |
| 8 |
250 |
36 |
1 |
28 |
250 |
36 |
1 |
28 |
|
| 9 |
350 |
6 |
1 |
29 |
|
|
|
|
|
| 10 |
350 |
9 |
2 |
30,31 |
|
|
|
|
|
| 11 |
350 |
12 |
8 |
12,32-34,62,81,87,109 |
350 |
12 |
2 |
17,32 |
10 |
| 12 |
350 |
24 |
16 |
1,9,36-46,51,59,69 |
350 |
24 |
6 |
9,36,37,51,59,69 |
22 |
| 13 |
350 |
48 |
2 |
47,60 |
350 |
48 |
2 |
11,47 |
4 |
| 14 |
350 |
120 |
1 |
48 |
350 |
120 |
1 |
48 |
2 |
| 15 |
400 |
12 |
1 |
49 |
400 |
12 |
1 |
33 |
2 |
| 16 |
400 |
24 |
1 |
50 |
400 |
24 |
1 |
39 |
2 |
| 17 |
490 |
12 |
1 |
55 |
490 |
12 |
1 |
55 |
2 |
| 18 |
500 |
12 |
3 |
52,53,65 |
500 |
12 |
4 |
6,22,62,135 |
7 |
| 19 |
500 |
48 |
3 |
10,73,80 |
500 |
48 |
2 |
73,80 |
5 |
| 20 |
500 |
60 |
1 |
54 |
500 |
60 |
2 |
54,67 |
3 |
| |
|
|
|
|
500 |
24 |
11 |
7,15,18,19,20,38,41,42,43,44,45 |
|
| |
|
|
|
|
600 |
24 |
1 |
50 |
|
Table 4: Relationship to define INR and IR from references by CD4+ T-cell count absolute number and the ART time.
From Figure 3, we can find that with regard to the definition of INR
and IR, because different references had different understanding
of INR, there was a phenomenon of data overlap. For example,
when ART time=24, the overlapping of CD4+ T-cell count absolute
number included 250,350 and 400.
In order to show the relationship more clearly, we processed the
definition of INR and IR in the references in the following way:
1. Deleted all symbols in the definition, including >, ≥, <, ≤.
2. For the definition of INR, if the same ART time corresponded
to multiple CD4+ T-cell count absolute number, the lowest
value was used.
3. For the definition of IR, if the same ART time corresponded
to multiple CD4+ T-cell count absolute number, the highest
value was used.
4. For data in a defined range, the minimum value of the range
was taken. For example, if the art time range was 6-12 months,
then the art time was taken as 6 months.
5. When the CD4+ T-cell count absolute number of INR and
IR were at the same ART time, since the value of INR usually
contained < or ≤, and the value of IR usually contained > or
≥, in order to show the difference between INR and IR, when
displaying the CD4+ T-cell count absolute number, we set the
CD4+ T-cell count absolute number of INR to -20, and the
CD4+ T-cell count absolute number of IR to +20.
6. If the CD4+ T-cell count absolute number corresponding to a
certain ART time was less than the value at the previous time
point but greater than the value at the next time point, this
point would be deleted.
Based on the above processing method, the relationship of INR
and IR displayed by CD4+ T-cell count absolute number and ART
time obtained from references was shown in Figure 4. From Figure
4, We found that between the two types of INR and IR, a line
drawn by the CD4+ T-cell count absolute number and ART time
may distinguish between INR and IR. This line may be a straight
line (the red line in Figure 4) or a curved line (the black line in Figure 4).
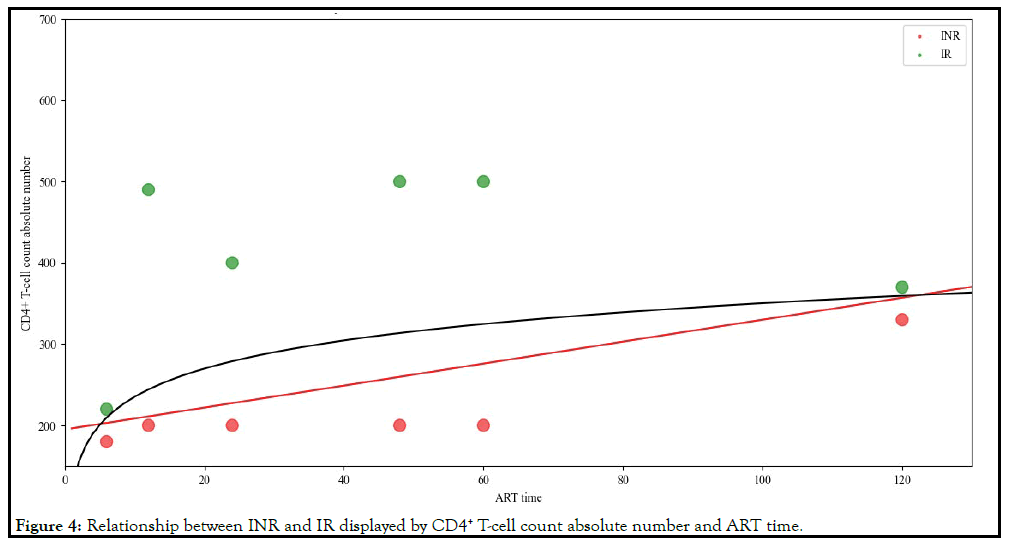
Figure 4: Relationship between INR and IR displayed by CD4+ T-cell count absolute number and ART time.
Classification result
Following the research in the previous section, we converted
the distinction between INR and IR into a supervised binary
classification problem. In order to facilitate the calculation of the
model, we proposed the following assumptions:
1. There was a certain mathematical relationship between CD4+ T-cell count absolute number and ART time. The model
established by this mathematical relationship can be used to
classify INR and IR.
2. Every doctor was scientific and credible for the diagnosis and
medication of INR.
3. Considering the serious harm of INR to the patient’s physical
condition, our definition of INR referred to the pessimistic
principle in management [137]. When a patient receives INR intervention treatment, but their CD4+ T-cell count was within the normal range, we still define it as INR.
Based on the above assumptions, we used the typical supervised
learning classification algorithm in machine learning to obtain a
model that can accurately determine INR through training. We
used the currently popular machine learning classification models
for modeling, including K-Nearest Neighbor (KNN), Least Absolute
Shrinkage and Selection Operator (Lasso), Ridge Regression,
Support Vector Machine(SVM), Decision Tree(DT), Gradient
Boosting Classifier(GBC), Logistic Regression(LR) and Multilayer
Perceptron(MLP). We used Cross-validation score (cross_val_score)
to determine the optimal classification model.
In order to avoid over-fitting and obtain the optimal classification
model, we adopted the shuffle-split cross-validation method, which
independently controlled the number of iterations in addition to
the size of the training set and the test set. The proportions of the
training set and the validation set were defined as 50% and 30%
respectively to ensure that a part of the data did not participate in
the training in each training time. The detailed modeling process
was shown in Part 2 of the supplement. Through modeling analysis,
the cross_val_score of each model and its corresponding optimal
parameters were shown in Table 5.
| No |
Model name |
Optimal hyperparameters |
Cross_val_score |
| 1 |
KNN |
'algorithm': 'ball_tree', 'leaf_size': 10, 'n_neighbors': 2, 'metric': 'chebyshev', 'weights': 'distance' |
0.9855 |
| 2 |
Lasso |
'alpha': 0.001, 'selection': 'cyclic', 'max_iter': 10000, 'tol': 0.0001 |
0.5949 |
| 3 |
Ridge |
'alpha': 0.001, 'solver': 'cholesky', 'max_iter': 1000, 'tol': 1e-06 |
0.5986 |
| 4 |
SVM |
'kernel': 'rbf', 'gamma': 10, 'C': 100, 'max_iter': 10000, 'tol': 0.0001 |
0.9911 |
| 5 |
DT |
'criterion': 'entropy', 'splitter': 'best', 'max_depth': 5, 'min_samples_leaf': 5 |
0.9461 |
| 6 |
GBC |
'learning_rate': 0.001, 'n_estimators': 90, 'max_depth': 5, 'min_samples_split': 800, 'min_samples_leaf': 60 |
0.7118 |
| 7 |
LR |
'solver': 'liblinear', 'penalty': 'l1', 'C': 10, 'max_iter': 1000, 'tol': 0.01 |
0.9669 |
| 8 |
MLP |
'hidden_layer_sizes': [20, 20], 'activation': 'identity', 'solver': 'lbfgs', 'alpha': 0.01, 'learning_rate': 'constant', 'max_iter': 10000, 'tol': 0.001 |
0.9675 |
Table 5: Cross-validation score of the supervised learning model and the corresponding optimal hyper parameters.
It can be seen from Table 5 that SVM had the best Crossvalidation
score. We can use the SVM model with the optimal
hyper parameters to define INR. The result of using SVM for
classification was shown in Figure 5.
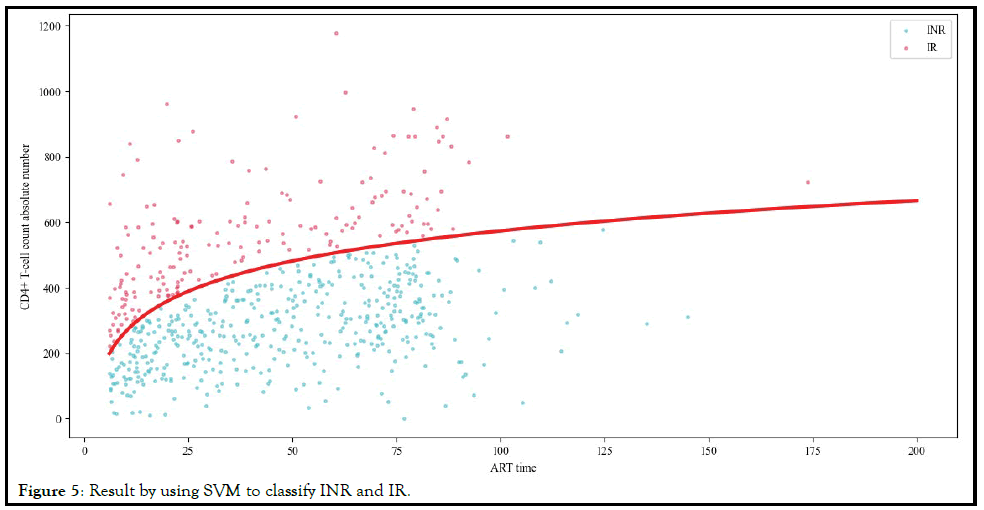
Figure 5: Result by using SVM to classify INR and IR.
Discussion
Reliability analysis of results and recommended INR
definition interval
Due to the influence of the data amount and outliers on the
credibility of the model, the results of the training model in this
paper were only valid for the current data. For example, in open
source data of reference [59], patients with CD4+ T-cell absolute
number=600 and ART time=106.8 were defined as INRs.
Normally, no matter what value of the ART time, patients with
CD4+ T-cell absolute number=600 should be regarded as the IRs,
but the author of this paper defined him as INRs. These data may
affect the credibility of the model.
At the same time, although the SVM model can assist in the
definition of INR, because this was a supervised learning model,
it was not convenient for clinicians to quickly determine whether
the patient was INRs or IRs. We also tried semi-supervised
learning algorithms and unsupervised learning algorithm, but
their classification accuracy and the interpretability were not as
good as supervised learning algorithms. The programming codes
of semi-supervised learning algorithm and unsupervised learning
algorithm was included in Part 3 and Part 4 of the appendix.
In order to facilitate scientific researchers and clinicians to quickly
and accurately defined the INR, through the supervised learning
classification model, we gave the recommended reference values of
the CD4+ T-cell absolute number in each time period of the ART,
as shown in Table 6. Based on our judgment, if the CD4+ T-cell
absolute number of a patient at a specific ART time was less than
the corresponding value in the table, it was considered that the
patient has a high probability of belonging to INRs.
| ART time |
CD4 |
ART time |
CD4 |
ART time |
CD4 |
ART time |
CD4 |
ART time |
CD4 |
| 6 |
199 |
17 |
338 |
28 |
404 |
39 |
448 |
50 |
482 |
| 7 |
219 |
18 |
345 |
29 |
409 |
40 |
452 |
51 |
484 |
| 8 |
237 |
19 |
353 |
30 |
413 |
41 |
455 |
52 |
487 |
| 9 |
253 |
20 |
359 |
31 |
418 |
42 |
458 |
53 |
489 |
| 10 |
267 |
21 |
366 |
32 |
422 |
43 |
461 |
54 |
492 |
| 11 |
280 |
22 |
372 |
33 |
426 |
44 |
465 |
55 |
494 |
| 12 |
291 |
23 |
378 |
34 |
430 |
45 |
468 |
56 |
497 |
| 13 |
302 |
24 |
384 |
35 |
434 |
46 |
470 |
57 |
499 |
| 14 |
312 |
25 |
389 |
36 |
438 |
47 |
473 |
58 |
501 |
| 15 |
321 |
26 |
394 |
37 |
441 |
48 |
476 |
59 |
504 |
| 16 |
330 |
27 |
399 |
38 |
445 |
49 |
479 |
60+ |
506 |
Note: CD4 was the abbreviation of CD4+ T-cell count absolute number.
Table 6: Criteria of CD4+ T-cell absolute number at each ART time for defining INR.
Considering that the SVM model was affected by the amount and
quality of data, we simplified Table 6 and proposed criteria for
defining INR by using CD4+ T-cell count absolute number and
ART Time, as shown in Table 7.
| ART time |
CD4 |
ART time |
CD4 |
| 6 |
200 |
30 |
410 |
| 9 |
250 |
36 |
440 |
| 12 |
300 |
42 |
460 |
| 18 |
350 |
48 |
480 |
| 24 |
380 |
60 |
500 |
Note: CD4 was the abbreviation of CD4+ T-cell count absolute number
Table 7: Simplified criteria of CD4+ T-cell absolute number at each ART time for defining INR.
Availability of other relevant features for defining INR
In our supervised learning classification model, due to the reasons
mentioned in the Features selection section, we did not use CD4+ T-cell count change number, CD4+ T-cell count growth rate, CD4/
CD8 ratio and VS time to define INR. This did not mean that we
thought these features were meaningless for defining INR. If these
data existed in actual use, on the basis of the recommended criteria
in this paper, we can regard the values of these features as adjuvant
standards to define INR. For example, the CD4/CD8 ratio less
than 1 could help define INR.
Conclusion
On the road to overcome AIDS, INR was still an important
research area. Finding the features and methods that accurately
define INR will help us understand INR more accurately and
discover the pathogenesis and interventions of INR. Through
systematic literature review, visualization analysis, and machine
learning modeling, we have discovered the consensus, features,
and supervised classification methods that could define INR. In
the future, we will collect more INR related data; introduce more
features and classification methods to obtain better ways to define
INR.
Ethics Approval and Consent to Participate
The study complied with the principles of the Declaration
of Helsinki and was approved by the Human Science Ethics
Committee of Chongqing Public Health Medical Center. The
Human Science Ethics Committee authorized the waiver of
informed consent based on the observational nature of this study,
and the ethics review approval document number: 2021-005-01-KY.
The data extracted from the electronic medical record database was
studied anonymously.
Acknowledgements
We would like to express our gratitude to all participants involved
in this study and the funder of the research.
Availability of Data and Material
The entire modeling process and source code could be obtained by
the corresponding author. The datasets used or analyzed during the
study were owned by Chongqing Public Health Medical Center.
Since the data includes sensitive patient data and involves some
patents under development, the data can only be shared after being
authorized by the corresponding author and the organization.
Funding
This study was supported by the Chongqing Science and Technology
Bureau Project ( cstc2019jscx-fxyd0298, cstc2020jscx-cylhX0001).
Consent for Publication
All authors have provided consent.
Competing Interests
Yong Shuai and Hemeng Peng contributed equally to this work.
References
- Zhao YL, Jiang Y, Ling H, Zhuang M. The mechanisms and related factors of poor immune reconstitution in HIV information individuals. Int J Immunol. 2020;43(6):603-609.
[Cross Ref]
- Silvar RB, Goios A, Kelly C, Teixeira P, João C, Horta A, et al. Definition of immunological non-response to antiretroviral therapy: A system review. J Acquir Immune Defic Syndr. 2019;82(5):452-461.
[Cross Ref] [Google Scholar] [PubMed]
- Lidia G, Camilla T, Giusi MB, Antonella AM, Giulia M. The absence of CD4+ T cell count recovery despite receipt of virologically suppressive highly active antiretroviral therapy: Clinical risk, immunological gaps, and therapeutic options. Clin Infect Dis. 2009;48(3):328-337.
[Cross Ref] [Google Scholar]
- Marta M, Eugnia N, Bonaventura C, Blanco J. Immunodiscordant responses to HAART-mechanisms and consequences. Expert Rev Clin Immunol. 2013;9(11):1135-1149.
[Cross Ref] [Google Scholar] [PubMed]
- Tobolowsky FA, Wada N, Martinez-Maza O, Magpantay L, Koletar L, Palella FJ, et al.Brief report: Circulating markers of fibrosis are associated with immune reconstitution status in HIV-infected men. PLoS One. 2018;13(1):e0191606.
[Cross Ref] [Google Scholar] [PubMed]
- Yang XD, Su B, Zhang X, Liu Y, Zhang T.Incomplete immune reconstitution in HIV/AIDS patients on antiretroviral therapy: Challenges of immunological non-responders. J Leukoc Biol. 2020;107(4):597-612.
[Cross Ref] [Google Scholar] [PubMed]
- Li CX, Li YY, He LP, Kou J, Bai JS, Liu J, et al.The predictive role of CD4+ cell count and CD4:CD8 ratio in immune reconstitution outcome among HIV/AIDS patients receiving antiretroviral therapy: An eight-year observation in China. BMC Immunol. 2019;20(1):31.
[Cross Ref] [Google Scholar] [PubMed]
- Li TS, Wang FS, Gao F. China guidellines for diagonosis and tratment of HIV/AIDS (2018). Med J Hosp. 2019;10(1):31-52.
[Cross Ref] [Google Scholar] [PubMed]
- Lu W, Feng YD, Jing FH, Han Y.Association between gut microbiota and CD4 recovery in HIV-1 infected patients. Front Micro Bio. 2018;9:1451.
[Cross Ref] [Google Scholar] [PubMed]
- Gibert CL. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. Health Human Serv. 2013;57:110-111.
[Cross Ref] [Google Scholar] [PubMed]
- Kong YX, Tian YF, Hao Y, Chong XJ, Xiao J, Yang D, et al. Two types of poor immunological responder showing distinct responses to long-term HAART. Int J Infect Dis. 2019;86:178-187.
[Cross Ref] [Google Scholar] [PubMed]
- Lebouché B, Yero A, Shi T, Farnos O, Singer J, Kema I, et al. Impact of extended release niacin on immune activation in HIV infected immunological non-responders on effective antiretroviral therapy. HIV Res Clin Pract. 2020;21(6):182-190.
[Cross Ref] [Google Scholar] [PubMed]
- Xia H, Wang Y, Sun HL, Gao LY, Cao Y, Zaongo SD, et al. Safety and efficacy of allogeneic natural killer cell immunotherapy on human immunodeficiency virus type 1 immunological non-responders: A brief report. Chin Med J. 2020;133(23):2803-2807.
[Cross Ref] [Google Scholar] [PubMed]
- Singh S, Toor JS, Sharma A, Arora SK. Signature genes associated with immunological non-responsiveness to anti-retroviral therapy in HIV-1 subtype-c infection. PLoS One. 2020;15(6):e0234270.
[Cross Ref] [Google Scholar] [PubMed]
- Marchetti G, Gori A, Casabianca A, Magnani M, Franzetti F, Clerici M, et al. Comparative analysis of T-cell turnover and homeostatic parameters in HIV-infected patients with discordant immune-virological responses to HAART. AIDS. 2006;20(13):1727-1736.
[Cross Ref] [Google Scholar] [PubMed]
- Bellistrì GM, Casabianca A, Merlini E, Orlandi C, Ferrario G, Meroni L, et al. Increased bone marrow interleukin-7 (IL-7)/IL-7R levels but reduced IL-7 responsiveness in HIV-positive patients lacking CD4+ gain on antiviral therapy. PLoS One. 2010;5(12):e15663.
[Cross Ref] [Google Scholar] [PubMed]
- Soria A, Guerini FR, Bandera A, Bolognesi E, Uglietti A, Fusco C , et al. KIR-HLA genotypes in HIV-infected patients lacking immunological recovery despite effective antiretroviral therapy. PLoS One. 2011;6(11):e27349.
[Cross Ref] [Google Scholar] [PubMed]
- Lelyveld SFL, Gras L, Kesselring A, Zhang SJ, Wolf FD Wensing AMJ, et al. Long-term complications in patients with poor immunological recovery despite virological successful HAART in Dutch ATHENA cohort. AIDS. 2012;26(4):465-474.
[Cross Ref] [Google Scholar] [PubMed]
- Marchetti G, Gazzola L, Trabattoni D, Bai F, Ancona G, Ferraris L, et al. Skewed T-cell maturation and function in HIV-infected patients failing CD4+ recovery upon long-term virologically suppressive HAART. AIDS. 2010 ;24(10):1455-1460.
[Cross Ref] [Google Scholar] [PubMed]
- Gaardbo JC, Hartling HJ, Ronit A, Springborg K, Gjerdrum LMR, Ralfkiær E, et al. Regulatory T cells in HIV- infected immunological nonresponders are increased in blood but depleted in lymphoid tissue and predict immunological reconstitution. J Acquir Immune Defic Syndr. 2014;66(4):349-357.
[Cross Ref] [Google Scholar] [PubMed]
- Loutfy MR, Genebat M, Moore D, Raboud J, Chan K, Antoniou T, et al. A CD4+ cell count , 200 cells per cubic millimeter at 2 Years after initiation of combination antiretroviral therapy is associated with increased mortality in HIV- infected individuals with viral suppression. J Acquir Immune Defic Syndr. 2010;55(4):451-459.
[Cross Ref] [Google Scholar] [PubMed]
- Molina-Pinelo S, Vallejo A, Díaz L, Soriano-Sarabia N, Ferrando-Martínez S, Resino S, et al. Premature immunosenescence in HIV-infected patients on highly active antiretroviral therapy with low-level CD4 T cell repopulation. J Antimicrob Chemother. 2009;64(3):579-588.
[Cross Ref] [Google Scholar] [PubMed]
- Pacheco YM, Jarrin I, Rosado I, Campins A A, Berenguer J, Iribarren J A, et al. Increased risk of non-AIDS- related events in HIV subjects with persistent low CD4 counts despite cART in the CoRIS cohort. Antiviral Research. 2015;117:69-74.
[Cross Ref] [Google Scholar] [PubMed]
- Pacheco YM, Jarrín I, Amo JD, Moreno S, Iribarren JA, Viciana P, et al. Risk factors, CD4 long-term evolution and mortality of HIV-infected patients who persistently maintain low CD4 counts, despite virological response to HAART. Curr HIV Res. 2009;7(6):612-619.
[Cross Ref] [Google Scholar] [PubMed]
- Rosado-Sánchez I, Herrero-Fernández I, Genebat M, Romero JD , Riera M, Podzamczer D, et al. HIV-infected subjects with poor CD4 T-Cell recovery despite effective therapy express high levels of OX40 and α4β7 on CD4 T-cells prior therapy initiation. Front Immunol. 2018;9:1673.
[Cross Ref] [Google Scholar] [PubMed]
- Rosado-Sánchez I, Herrero-Fernández I, Álvarez-Ríos AI, Genebat M, Abad-Carrillo MA, Ruiz-Mateos E, et al. A lower baseline CD4/CD8 T-cell ratio is independently associated with immunodiscordant response to antiretroviral therapy in HIV-infected subjects. Antimicrob. Agents Chemother. 2017;61(8):e00605-17.
[Cross Ref] [Google Scholar]
- Rosado-Sánchez I, Jarrín I, Pozo-Balado MM, Pablo-Bernal RS, Herrero-Fernández I, Alvarez-Ríos AI, et al. Higher levels of IL-6, CD4 turnover and Treg frequency are already present before cART in HIV-infected subjects with later low CD4 recovery. Antiviral Res. 2017;142:76-82.
[Cross Ref] [Google Scholar] [PubMed]
- Rodríguez-Gallego E, Gómez J, Pacheco YM, Peraire J, Viladés C, Beltrán-Debón R, et al. A baseline metabolomic signature is associated with immunological CD4+ T-cell recovery after 36 months of antiretroviral therapy in HIV-infected patients. AIDS. 2018;32(5):565-573.
[Cross Ref] [Google Scholar] [PubMed]
- Hunt PW, Martin JN, Sinclair E, Epling L, Teague J, Jacobson MA, et al. Valganciclovir reduces T cell activation in HIV-infected individuals with incomplete CD4+ T cell recovery on antiretroviral therapy. J Infect Dis. 2011;203(10):1474-1483.
[Cross Ref] [Google Scholar] [PubMed]
- Minami R, Takahama S, Kaku Y, Yamamoto M. Addition of maraviroc to antiretroviral therapy decreased interferon-γ mRNA in the CD4+ T cells of patients with suboptimal CD4+. J Infect Chemother. 2017;23(1):29-34.
[Cross Ref] [Google Scholar] [PubMed]
- Routy JP, Angel JB, Patel M, Kanagaratham C, Radzioch D, Kema I, et al. Assessment of chloroquine as a modulator of immune activation to improve CD4 recovery in immune nonresponding HIV-infected patients receiving antiretroviral therapy. HIV Med. 2015;16(1):48-56.
[Cross Ref] [Google Scholar] [Pubmed]
- Zhang FD, Sun MY, Sun JJ, Guan LQ, Wang J, Lu H, et al. The risk factors for suboptimal CD4 recovery in HIV infected population: An observational and retrospective study in Shanghai, China. Biosci Trends. 2015;9(5):335-341.
[Cross Ref] [Google Scholar] [Pubmed]
- Falster K, Petoumenos K, Chuah J, Mijch A, Mulhall B, Kelly M et al. Poor baseline immune function predicts an incomplete immune response to combination antiretroviral treatment despite sustained viral suppression. J Acquir Immune Defic Syndr. 2009;50(3):307-313.
[Cross Ref] [Google Scholar] [Pubmed]
- Casotti JAS, Passos LN, Oliveira FJP, Cerutti C. Prevalence of discordant immunologic and virologic responses in patients with AIDS under antiretroviral therapy in a specialized care center in Brazil. Rev Inst Med Trop Sao Paulo. 2011;53(6):301-307.
[Cross Ref] [Google Scholar]
- Shmagel NG, Shmagel KV, Saidakova EV, Korolevskaya LB, Chereshnev VA. Discordant response of CD4+ T cells to antiretroviral therapy in HIV-infected patients coinfected with hepatitis C virus is accompanied by increased liver damage. Dokl Biochem Biophys. 2015;465:358-360.
[Cross Ref] [Google Scholar] [Pubmed]
- Saidakova EV, Korolevskaya LB, Shmagel NG, Shmagel KV, Chereshnev VA. The role of interleukin 7 and its cell receptor in a poor recovery of CD4+ T cells in HIV-infected patients receiving antiretroviral therapy. Dokl Biol Sci. 2014;458:313-315.
[Cross Ref] [Google Scholar] [Pubmed]
- Sennepin A, Baychelier F, Guihot A, Nel I, Fang RHT, Calin R, et al. NKp44L expression on CD4+ T cells is associated with impaired immunological recovery in HIV-infected patients under highly active antiretroviral therapy. AIDS. 2013;27(12):1857-1866.
[Cross Ref] [Google Scholar] [Pubmed]
- Giuliani E, Vassena L, Cesare S, Malagnino V, Desimio MG, Andreoni M, et al. NK cells of HIV-1-infected patients with poor CD4+ T-cell reconstitution despite suppressive HAART show reduced IFN-γ production and high frequency of autoreactive CD56 bright cells. Immunol Lett. 2017;190(12):185-193.
[Cross Ref] [Google Scholar]
- Massanella M, Negredo E, Pérez-Alvarez N, Puig J, Ruiz-Hernández R, Bofill M, et al. CD4 T-cell hyperactivation and susceptibility to cell death determine poor CD4 T-cell recovery during suppressive HAART. AIDS. 2010;24(7):959-968.
[Cross Ref] [Google Scholar] [Pubmed]
- Negredo E, Massanella M, Puertas MC, Buzón MJ, Puig J, Pérez-Alvárez N, et al. Early but limited effects of raltegravir intensification on CD4 T cell reconstitution in HIV-infected patients with an immunodiscordant response to antiretroviral therapy. J. Antimicrob. Chemother. 2013;68(10):2358-2362.
[Cross Ref] [Google Scholar]
- Shive CL, Clagett B, McCausland MR, Mudd JC, Funderburg NT, Freeman ML et al. Inflammation perturbs the IL-7 Axis, promoting senescence and exhaustion that broadly characterize immune failure in treated HIV infection. J Acquir Immune Defic Syndr. 2016;71(5):483-492.
[Cross Ref] [Google Scholar] [Pubmed]
- Lee SC, Chua LL, Yap SH, Khang TF, Leng CY, Azwa RIR, et al. Enrichment of gut-derived Fusobacterium is associated with suboptimal immune recovery in HIV-infected individuals. Sci Report. 2018;8(1):14277.
[Cross Ref] [Google Scholar] [Pubmed]
- Bandera A, Masetti M, Fabbiani M, Biasin M, Muscatello A, Squillace N et al. The NLRP3 inflammasome is upregulated in HIV-infected antiretroviral therapy-treated individuals with defective immune recovery. Frontiers Immun. 2018;9:214.
[Cross Ref] [Google Scholar] [Pubmed]
- Benedetto ID, Masetti M, Fabbiani M, Biasin M , Muscatello A, Squillace N, et al. Higher levels of peripheral Th17 T CD4+ cells are associated with immunological non response in HIV-infected patients under effective ART. J Acquir Immune Defic Syndr. 2018;77(5):e45-e47.
[Cross Ref] [Google Scholar] [Pubmed]
- Wójcik-Cichy K, Piekarska A, Jabłonowska E. Intestinal barrier impairment and immune activation in HIV-infected advanced late presenters are not dependent on CD4 recovery. Arch Immunol Ther Exp. 2018;66(4):321-327.
[Cross Ref] [Google Scholar] [Pubmed]
- Logerot S, Rancez M, Muylder BC, Figueiredo-Morgado S, Sandrad R, Giuseppe T, et al. HIV reservoir dynamics in HAART-treated poor immunological responder patients under IL-7 therapy. AIDS. 2018;32(6):715-720.
[Cross Ref] [Google Scholar] [Pubmed]
- Gunda DW, Kilonzo SB, Kamugisha E, Rauya EZ, Mpondo BC. Prevalence and risk factors of poor immune recovery among adult HIV patients attending care and treatment center in northwestern Tanzania following the use of highly active antiretroviral therapy: A retrospective study. BMC Res Notes. 2017;10(1):197.
[Cross Ref] [Google Scholar] [Pubmed]
- Gómez-Mora E, Massanella M, García E, Giles D, Bernadó M, Urrea V, et al. Elevated humoral response to cytomegalovirus in HIV-infected individuals with poor CD4+ T-cell immune recovery. PLoS One. 2017;12(9):e0184433.
[Cross Ref] [Google Scholar] [Pubmed]
- Thiébaut R, Jarne A, Routy JP, Sereti I, Fischl M, Ive P, et al. Repeated cycles of recombinant human interleukin 7 in HIV-infected patients with low CD4 T-cell reconstitution on antiretroviral therapy. Clin Infect Dis. 2016;62(9):1178-1185.
[Cross Ref] [Google Scholar] [Pubmed]
- Stiksrud B, Lorvik KB, Kvale D, Mollnes TE, Ueland PM, Trøseid M, et al. Plasma IP-10 is increased in immunological nonresponders and associated with activated regulatory T cells and persisting low CD4 counts. J Acquir Immune Defic Syndr. 2016;73(2):138-148.
[Cross Ref] [Google Scholar] [Pubmed]
- Saison J, Ferry T, Demaret J, Boulch DM, Venet F, Perpoint T,et al. Association between discordant immunological response to highly active anti-retroviral therapy, regulatory T cell percentage, immune cell activation and very low-level viraemia in HIV-infected patients. Clin Exp Immunol. 2014;176(3):401-409.
[Cross Ref] [Google Scholar] [Pubmed]
- Tanaskovic S, Fernandez S, French MA, Price RI, et al. Thymic tissue is not evident on high-resolution computed tomography and [18F] fluoro-deoxy-glucose positron emission tomography scans of aviraemic HIV patients with poor recovery of CD4+ T cells. AIDS. 2011;25(9):1235-1237.
[Cross Ref] [Google Scholar] [Pubmed]
- Horta A, Nobrega C, Amorim-Machado P, Coutinho-Teixeira V, Barreira-Silva P, Boavida S, et al. Poor immune reconstitution in HIV-infected patients associates with high percentage of regulatory CD4+ T cells. PLoS One. 2013;8(2):e57336.
[Cross Ref] [Google Scholar] [Pubmed]
- Tasca KI, Correa CR, Caleffi JT, Mendes MB, Gatto M, Manfio VM, et al. Asymptomatic HIV people present different profiles of sCD14, sRAGE, DNA damage, and vitamins, according to the use of cART and CD4+ T cell restoration. J Immunol Res. 2018;53:1-11.
[Cross Ref] [Google Scholar]
- Fernandez S, Tanaskovic S, Helbig K, Rajasuriar R, Kramski M, Murray JM et al. CD4+ T-cell deficiency in HIV patients responding to antiretroviral therapy is associated with increased expression of interferon-stimulated genes in CD4+ T cells. J Infect Dis. 2011;204(12):1927-1935.
[Cross Ref] [Google Scholar] [Pubmed]
- Zhu QD, Liang G, Chen W, Wu FY, Lu XC. Clinical analysis of endocrine metabolism in 50 patients with poor immune reconstitution after ART. Frontiers Medicine. 2018; 20:97-98.
[Cross Ref] [Google Scholar]
- Lichtenstein KA, Armon C, Nagabhushanam V, Efaw BJ, Frazer-Abel A, Hiserote ME, et al. A pilot study to assess inflammatory biomarker changes when raltegravir is added to a virologically suppressive HAART regimen in HIV-1-infected patients with limited immunological responses. Antivir Ther. 2012;17(7):1301-1309.
[Cross Ref] [Google Scholar]
- Wilkin TJ, Lalama CM, McKinnon J, Gandhi RT, Lin N, Landay A, et al. A pilot trial of adding maraviroc to suppressive antiretroviral therapy for suboptimal CD4+ T-cell recovery despite sustained virologic suppression: ACTG A5256. J Infect Dis. 2012;206(4):534-42.
[Cross Ref] [Google Scholar] [Pubmed]
- Lu DF, Zhang JB, Wang YX, Geng ST, Zhang Z, Xu Y et al. Association between CD4+ T cell counts and gut microbiota and serum cytokines levels in HIV-infected immunological non-responders. BMC Infect Dis 2021;21(1):742.
[Cross Ref] [Google Scholar] [Pubmed]
- Lilian RR, Davies N, Gilbert L, McIntyre JA, Struthers HE, Rees K, et al. CD4 testing after initiation of antiretroviral therapy: Analysis of routine data from the South African HIV programme. South Afr J HIV Med 2020;21(1):1165.
[Cross Ref] [Google Scholar] [Pubmed]
- Zoufaly A, Heiden MAD, Kollan C, Bogner JR, Fätkenheuer G, Wasmuth JC, et al. Clinical outcome of HIV- infected patients with discordant virological and immunological response to antiretroviral therapy. J Infect Dis. 2011;203(3):364-371.
[Cross Ref] [Google Scholar] [Pubmed]
- Zhang LX, Song JW, Zhang C, Fan X, Huang H, Xu R , et al. Dynamics of HIV reservoir decay and naive CD4 T-cell recovery between immune non-responders and complete responders on long-term antiretroviral treatment. J Clin Immunol. 2021; 229:108773.
[Cross Ref] [Google Scholar] [Pubmed]
- Magen E, Elbirt D, Agmon-Levin N, Mishal J, Sthoeger Z. Eradication of Helicobacter pylori can facilitate immune reconstitution in HIV-1-infected immunological non-responders. Int J Infec Dis. 2010;14(4):e322-e327.
[Cross Ref] [Google Scholar]
- Merlini E, Bai F, Bellistrì GM, Tincati C, Monforte AD, Marchetti G. Evidence for polymicrobic flora translocating in peripheral blood of HIV-infected patients with poor immune response to antiretroviral therapy. PLoS One. 2011;6(4):e18580.
[CrossRef] [Google Scholar] [Pubmed]
- Julg B, Poole D, Ghebremichael M, Castilla C, Altfeld M, Sunpath H, et al. Factors predicting discordant virological and immunological responses to antiretroviral therapy in HIV-1 clade C infected Zulu/Xhosa in South Africa. PLoS One. 2012;7(2):e31161.
[CrossRef] [Google Scholar] [Pubmed]
- Raffi F, Moing VL, Assuied A, Habak S, Spire B, Cazanave C, et al. Failure to achieve immunological recovery in HIV-infected patients with clinical and virological success after 10 years of combined ART: Role of treatment course. J Anti Chem. 2017;72(1):240-245.
[CrossRef] [Google Scholar] [Pubmed]
- Nakanjako D, Kiragga AN, Musick BS, Yiannoutsos CT, Kaloustian KW, Diero L, et al. Frequency and impact of suboptimal immune recovery on first-line antiretroviral therapy within the international epidemiologic databases to evaluate AIDS in East Africa. AIDS. 2016;30(12):1913-1922.
[CrossRef] [Google Scholar] [Pubmed]
- Restrepo C, Gutierrez-Rivas M, Pacheco YM, García M, Blanco J, Medrano LM, et al. Genetic variation in CCR2 and CXCL12 genes impacts on CD4 restoration in patients initiating cART with advanced immunesupression. PLoS One. 2019;14(3):e0214421.
[CrossRef] [Google Scholar] [Pubmed]
- Melliez H, Prost M, Behal H, Neveux N, Benoist JF, Kim I, et al. Hypervitaminosis A is associated with immunological non-response in HIV-1-infected adults a case-control study. Euro J Clin Micro Infec Dis. 2020;39(11):2091-2098.
[CrossRef] [Google Scholar] [Pubmed]
- Meissner EG, Chung DJ, Tsao B, Haas DW, Utay NS. IFNL4 genotype does not associate with CD4 T-cell recovery in people living with human immunodeficiency virus. AIDS Res Hum Retroviruses. 2021;37(3):184-188.
[CrossRef] [Google Scholar] [Pubmed]
- Anude CJ, Eze E, Onyegbutulem HC, Charurat M, Etiebet M, Ajayi S, et al. Immuno-virologic outcomes and immuno-virologic discordance among adults alive and on anti-retroviral therapy at 12 months in Nigeria. BMC Infect Dis. 2013;13:113.
[CrossRef] [Google Scholar]
- Hermans SM, Kiragga AN, Schaefer P, Kambugu A, Hoepelman AIM, Manabe YC, et al. Incident tuberculosis during antiretroviral therapy contributes to suboptimal immune recon- stitution in a large urban HIV clinic in sub-saharan africa. PLoS One. 2010;5(5):e10527.
[CrossRef] [Google Scholar] [Pubmed]
- Zhang QY, Zhang X, Su B, Liu LF, Yang X, Tang B et al. Increased early activation of CD56dimCD16dim:- natural killer cells in immunological non-responders correlates with CD4+ T-cell recovery. Chin Med J. 2020;133(24):2928-2939.
[CrossRef] [Google Scholar] [Pubmed]
- Byakwaga H, Kell M, Purcell DFJ, French MA, Amin J, Lewin SR,et al. Intensification of antiretroviral therapy with raltegravir or addition of hyperimmune bovine colostrum in HIV-infected patients with suboptimal CD4+ T-cell response: a randomized controlled trial. J Infect Dis. 2011;204(10):1532-1540.
[CrossRef] [Google Scholar] [Pubmed]
- Tincati C, Merlini E, Braidotti P, Ancona G, Savi F, Tosi D,et al. Impaired gut junctional complexes feature late-treated individuals with suboptimal CD4+ T-cell recovery upon virologically suppressive combination antiretroviral therapy. AIDS. 2016;30(7):991-1003.
[CrossRef] [Google Scholar] [Pubmed]
- Asdamongkol N, Phanachet P, Sungkanuparph S. Low plasma zinc levels and immunological responses to zinc supplementation in HIV- infected patients with immunological discordance after antiretroviral therapy. Jpn J Infect Dis. 2013;66(6):469-474.
[CrossRef] [Google Scholar] [Pubmed]
- Rusconi S, Vitiello P, Adorni F, Colella E, Focà E, Capetti A, et al. Maraviroc as intensification strategy in HIV-1 positive patients with deficient immunological response: An Italian randomized clinical trial. PLoS One. 20134;8(11):e80157.
[CrossRef] [Google Scholar] [Pubmed]
- Lelyveld SFL, Drylewicz J, Krikke M, Veel EM, Otto SA, Richter C, et al. Maraviroc intensification of cART in patients with suboptimal immunological recovery: A 48-Week, placebo-controlled randomized trial. PLoS One. 2015;10(7):e0132430.
[CrossRef] [Google Scholar] [Pubmed]
- Asmelash A, Zheng Y, Kaloustian KW, Shaffer D, Sawe F, Ogwu A et al. Predictors of suboptimal CD4 response among women achieving virologic suppression in a randomized antiretroviral treatment trial, Africa. BMC Infect Dis. 2014;14:331.
[CrossRef] [Google Scholar] [Pubmed]
- Kim KH, Yi JY, Lee SH. The CD4 slope can be a predictor of immunologic recovery in advanced HIV patients: A case- control study. Korean J Intern Med. 2015;30(5):705-713.
[CrossRef] [Google Scholar] [Pubmed]
- Boucau J, Madouasse J, Kourjian G, Carlin CS, Wambua D, Berberich MJ, et al. The activation state of CD4 T cells alters cellular peptidase activities, HIV antigen processing, and mhc class i presentation in a sequence-dependent manner. J Immunol. 2019;202(10):2856-2872.
[CrossRef] [Google Scholar] [Pubmed]
- Somsouk M, Dunham RM, Cohen M, Albright R, Abdel-Mohsen M, Liegler T et al. The immunologic effects of mesalamine in treated HIV-infected individuals with incomplete CD4+ T cell recovery: a randomized crossover trial. PLoS One. 2014;9(12):e116306.
[CrossRef] [Google Scholar] [Pubmed]
- Li TS, Xie J, Li YJ, Jean-Pierre Routy, Li Y, Han Y et al. Tripterygium wilfordii Hook F extract in cART-treated HIV patients with poor immune response: A pilot study to assess its immunomodulatory effects and safety. HIV Clin Trials. 2015;16(2):49-56.
[CrossRef] [Google Scholar] [Pubmed]
- Cillo AR, Hilldorfer BB, Lalama CM, McKinnon JE, Coombs RW, Tenorio AR, et al. Virologic and immunologic effects of adding maraviroc to suppressive antiretroviral therapy in individuals with suboptimal CD4+ T-cell recovery. AIDS. 2015;29(16):2121-2129.
[CrossRef] [Google Scholar] [Pubmed]
- Su C, Peng X, Liu JJ. Clinical observation of Peiyuan capsule combined with HAART on immune reconstruction failure of AIDS. Chinese J AIDS STD. 2020;26(1):19:22.
[CrossRef] [Google Scholar]
- Tang BB, Li ZM, Gao HB ,Luo LJ. Meta-analysis of traditional chinese medicine combined with haart intervention for poor imune reconstitution in AIIDS patients. ACTA Chinese Med. 2018;33(243):1375-1379.
[CrossRef] [Google Scholar]
- Zhao Q ,Luo XQ, Xu K, Rong YX. A clinical study on self-made Jianpi Yiqi Formula combined with HARRT on promoting immune reconstruction of spleen deficiency and dampness syndrome in patients with AIDS. Clinical J Chinese Med. 2018; 10(30): 102-105.
[CrossRef] [Google Scholar]
- Meyer-Myklestad MH, Medhus AW, Lorvik KB, Seljeflot I, Hansen SH, Holm K, et al. Human immunodeficiency virus-infected immunological nonresponders have colon-restricted gut mucosal immune dysfunction. J Infect Dis. 2022;225(4):661-674.
[CrossRef] [Google Scholar] [Pubmed]
- Briceño O, Chávez-Torres M, Peralta-Prado A, Garrido-Rodríguez D, Romero-Mora K, Pinto-Cardoso S, et al. Associations between recent thymic emigrants and CD4+ T-cell recovery after short-term antiretroviral therapy initiation. AIDS. 2020; 34(4): 501-511.
[CrossRef] [Google Scholar] [Pubmed]
- Valdés-Ferrer SI, Crispín JC, Belaunzarán-Zamudio PF, Rodríguez-Osorio CA, Cacho-Díaz B, Alcocer-Varela J, et al. Add-on Pyridostigmine Enhances CD4+ T-Cell Recovery in HIV-1-Infected Immunological Non-Responders-A Proof-of-Concept Study. Front Immunol. 2017;8:1301.
[CrossRef] [Google Scholar] [Pubmed]
- Coelho AVC, Moura RR, Guimaraes RF, Brandao LAC, Crovella S. Antiretroviral therapy immunologic non-response in a Brazilian population: Association study using pharmaco- and immunogenetic markers. Braz J Infect Dis. 2018;22(5):392-401.
[CrossRef] [Google Scholar] [Pubmed]
- Meyer-Myklestad MH, Medhus AW, Lorvik KB, Seljeflot I, Hansen SH, Holmet K, et al. HIV-infected immunological non-responders have colon-restricted gut mucosal immune dysfunction. J Infect Dis. 2022;225(4):661-674.
[CrossRef] [Google Scholar]
- Shete A, Dhayarkar S, Sangale S, Medhe U, Panchal N, Rahane G, et al. Incomplete functional T-cell reconstitution in immunological non-responders at one year after initiation of antiretroviral therapy possibly predisposes them to infectious diseases. Int J Infect Dis. 2019;81:114-122.
[CrossRef] [Google Scholar] [Pubmed]
- Jacobson JM, Wang HY, Bordi R, Zheng L, Gross BH, Landay AL, et al. A randomized controlled trial of palifermin (recombinant human keratinocyte growth factor) for the treatment of inadequate CD4+ T-lymphocyte recovery in patients with HIV-1 infection on antiretroviral therapy. J Acquir Immune Defic Syndr. 2014;66(4):399-406.
[CrossRef] [Google Scholar] [Pubmed]
- Erikstrup C, Kronborg G, Lohse N, Ostrowski SR, Gerstoft J, Ullum H. T-Cell dysfunction in HIV-1–infected patients with impaired recovery of CD4 cells despite suppression of viral replication. J Acquir Immune Defic Syndr. 2010;53(3):303-310.
[CrossRef] [Google Scholar] [Pubmed]
- Engsig FN, Zangerle R, Katsarou O, Dabis F, Reiss P, Gill J et al. Long-term mortality in HIV-positive individuals virally suppressed for 3 years with incomplete CD4 recovery. Clin Infect Dis. 2014;58(9):1312-1321.
[CrossRef] [Google Scholar] [Pubmed]
- Stepanyuk O, Chiang TS, Dever LL, Paez SL, Smith SM, Perez G, et al. Impact of adding maraviroc to antiretroviral regimens in patients with full viral suppression but impaired CD4 recovery. AIDS. 2009;23(14):1911-1913.
[CrossRef] [Google Scholar] [Pubmed]
- Méndez-Lagares G, Pozo-Balado MM, Genebat M, Pergañeda AG, Leal M, Pacheco YM. Severe immune dysregulation affects CD4+CD25hiFoxP3+ regulatory T cells in HIV-infected patients with low-level CD4 T-cell repopulation despite suppressive highly active antiretroviral therapy. J Infect Dis. 2012;205(10):1501-1509.
[CrossRef] [Google Scholar] [Pubmed]
- Méndez-Lagares G, García-Pergañeda A, Pozo-Balado MM, Genebat M, Ruiz-Mateos E, García MG, et al. Differential alterations of the CD4 and CD8 T cell subsets in HIV-infected patients on highly active antiretroviral therapy with low CD4 T cell restoration. J Antimicrob Chemother. 2012 May;67(5):1228-1237.
[CrossRef] [Google Scholar] [Pubmed]
- Hunt PW, Shulman NS, Hayes TL, Dahl V, Somsouk M, Funderburg NT, et al. The immunologic effects of maraviroc intensification in treated HIV-infected individuals with incomplete CD4+ T-cell recovery: A randomized trial. Blood. 2013;121(23):4635-4646.
[CrossRef] [Google Scholar] [Pubmed]
- Casotti JAS, Passos LN, Oliveira FJP, Crispim CJ. Factors associated with paradoxical immune response to antiretroviral therapy in HIV infected patients: A case control study. BMC Infect Dis. 2011;11:306.
[Google Scholar] [Cross Ref] [Pubmed]
- Hatano H, Hayes TL, Dahl V, Sinclair E, Lee TH, Hoh R, et al. A randomized, controlled trial of raltegravir intensification in antiretroviral-treated, HIV-infected patients with a suboptimal CD4+ T cell response. J Infect Dis. 2011;203(7):960-968.
[Google Scholar] [Cross Ref] [Pubmed]
- Massanella M, Gómez-Mora E, Carrillo J, Curriu M. Increased ex vivo cell death of central memory CD4 T cells in treated HIV infected individuals with unsatisfactory immune recovery. J Translational Medicine. 2015;13:230.
[Google Scholar] [Cross Ref]
- Massanella M, Negredo E, Puig J, Puertas MC, Buzón MJ, Pérez-Álvarez N, et al. Raltegravir intensification shows differing effects on CD8 and CD4 T cells in HIV-infected HAART-suppressed individuals with poor CD4 T-cell recovery. AIDS. 2012;26(18):2285-93.
[Google Scholar] [Cross Ref] [Pubmed]
- Negredo E, Massanella M, Puig J, Pérez-Alvarez N, Gallego-Escuredo JM, Villarroya J, et al. Nadir CD4 T cell count as predictor and high CD4 T cell intrinsic apoptosis as final mechanism of poor CD4 T cell recovery in virologically suppressed HIV-infected patients: Clinical implications. Clin Infect Dis. 2010;50(9):1300-1308.
[Google Scholar] [Cross Ref] [Pubmed]
- Girard A, Vergnon-Miszczycha D, Depincé-Berger AE, Roblin X, Lutch F, Lambert C, et al. A high rate of β7+ gut-homing lymphocytes in hiv-infected immunological nonresponders is associated with poor CD4 T-cell recovery during suppressive HAART. J Acquir Immune Defic Syndr. 2016;72(3):259-265.
[Google Scholar] [Cross Ref] [Pubmed]
- Tuboi SH, Pacheco AG, Harrison LH, Stone RA, May M, Brinkhof MWG, et al. Mortality associated with discordant responses to antiretroviral therapy in resource-constrained settings. J Acquir Immune Defic Syndr. 2010;53(1):70-77.
[Google Scholar] [Cross Ref] [Pubmed]
- Batista G, Buvé A, Gueye NFN, Manga NM, DiopMN, Ndiaye K, et al. Initial suboptimal CD4 reconstitution with antiretroviral therapy despite full viral suppression in a cohort of HIV-infected patients in Senegal. Med Mal Infect. 2015;45(6):199-206.
[Google Scholar] [Cross Ref] [Pubmed]
- Griensven JV, Zachariah R, Rasschaert F, Reid T, Freya MD, Tony R. Discordant immunologic and virologic responses to antiretroviral therapy and associated mortality in a large treatment program in Rwanda. J Acq Immune Def Synd. 2009;50(5):556-8.
[Google Scholar] [Cross Ref]
- El-Beeli M, Al-Mahrooqi SH, Youssef RM, Zadjali F, Balkhair A, Said Al-Balushi M, et al. HLA-A68 and HLA-B15 alleles correlate with poor immune response among AIDS patients on combined antiretroviral therapy. Hum Immunol. 2016;77(6):490-497.
[Google Scholar] [Cross Ref] [Pubmed]
- Kayigamba FR, Franke MF, Bakker MI, Rodriguez CA, Bagiruwigize E, Wnm Wit F, et al. Discordant treatment responses to combination antiretroviral therapy in rwanda: A prospective cohort study. PLoS One. 2016;11(7):e0159446.
[Google Scholar] [Cross Ref] [Pubmed]
- Su QJ, Li YZ, Liang FL, Xiao J, Deng X. Polyactin A increases CD4+ T-cell counts in HIV-infected individuals with insufficient immunologic response to highly active antiretroviral therapy. Int J STD & AIDS. 2014;25(1):24-28.
[Google Scholar] [Cross Ref]
- Gilson RJC, Man SL, Copas A, Rider A, Forsyth S, Hill T, et al. Discordant responses on starting highly active antiretroviral therapy: suboptimal CD4 increases despite early viral suppression in the UK Collaborative HIV Cohort (UK CHIC) Study. HIV Med. 2010;11(2):152-160.
[Google Scholar] [Cross Ref] [Pubmed]
- Onen NF, Overton ET, Presti R, Blair C, Powderly WG, Mondy K. Sub-optimal CD4 recovery on long-term suppressive highly active antiretroviral therapy is associated with favourable outcome. HIV Med. 2009;10(7):439-446.
[Google Scholar] [Cross Ref] [Pubmed]
- Woelk CH, Beliakova-Bethell N, Goicoechea M, Zhao YD, Du P, Rought SE, et al. Gene expression before haart initiation predicts hiv-infected individuals at risk of poor CD4+ t-cell recovery. AIDS. 2010;24(2):217-222.
[Google Scholar] [Cross Ref] [Pubmed]
- Rallón N, Sempere-Ortells JM, Soriano V, Benito JM. Central memory CD4 T cells are associated with incomplete restoration of the CD4 T cell pool after treatment-induced long-term undetectable HIV viraemia. J Antimicrob Chemother. 2013;68(11):2616-2625
[Google Scholar] [Cross Ref] [Pubmed]
- Mavigner M, Delobel P, Cazabat M, Dubois M, L'Faqihi-Olive FE, Raymond S, et al. HIV-1 residual viremia correlates with persistent T-cell activation in poor immunological responders to combination antiretroviral therapy. PLoS One. 2009;4(10):e7658.
[Google Scholar] [Cross Ref]
- Mupfumi L, Moyo S, Molebatsi K, Thami PK, Anderson M, Mogashoa T, et al. Immunological non-response and low hemoglobin levels are predictors of incident tuberculosis among HIV-infected individuals on Truvada-based therapy in Bot-swana. PLoS One. 2018;13(1):e0192030.
[Google Scholar] [Cross Ref] [Pubmed]
- Nakanjako D, Ssewanyana I, Mayanja-Kizza H, Kiragga A, Colebunders R, Manabe YC, et al. High T-cell immune activation and immune exhaustion among individuals with suboptimal CD4 recovery after 4 years of antiretroviral therapy in an African cohort. BMC Infect Dis. 2011;11:43.
[Google Scholar] [Cross Ref] [Pubmed]
- Bayigga L, Nabatanzi R, Sekiziyivu PN, Mayanja-Kizza H, Kamya MR, Kambugu A, et al. High CD56++CD16-natural killer (NK) cells among suboptimal immune responders after four years of suppressive antiretroviral therapy in an African adult HIV treatment cohort. BMC Immunol. 2014;15:2.
[Google Scholar] [Cross Ref] [Pubmed]
- Nakanjako D, Ssewanyana I, Nabatanzi R, Kiragga A, Kamya MR, Cao H, et al. Impaired T-cell proliferation among HAART-treated adults with suboptimal CD4 recovery in an African cohort. BMC Immunol. 2013;14:26.
[Google Scholar] [Cross Ref] [Pubmed]
- Nakanjako D, Ssinabulya I, Nabatanzi R, Bayigga L, Kiragga A, Joloba M, et al. Atorvastatin reduces T-cell activation and exhaustion among HIV-infected cART-treated suboptimal immune responders in Uganda: a randomised crossover placebo-controlled trial. Trop Med Int Health. 2015;20(3):380-390.
[Google Scholar] [Cross Ref] [Pubmed]
- Mingbunjerdsuk P, Asdamongkol N, Sungkanuparph S. Factors associated with immunological discordance in HIV-infected patients receiving antiretroviral therapy with complete viral suppression in a resource-limited setting. Jpn J Infect Dis. 2015;68(4):301-304.
[Google Scholar] [Cross Ref] [Pubmed]
- Takuva S, Maskew M, Brennan AT, Long L, Sanne I, Fox MP. Poor CD4 recovery and risk of subsequent progression to AIDS or death despite viral suppression in a South African cohort. J Int AIDS Soc. 2014;17(1):18651.
[Google Scholar] [Cross Ref] [Pubmed]
- Darraj M, Shafer LA, Chan S, Kasper K, Keynan Y. Rapid CD4 decline prior to antiretroviral therapy predicts subsequent failure to reconstitute despite HIV viral suppression. J Infect Public Health. 2018;11(2):265-269.
[Google Scholar] [Cross Ref] [Pubmed]
- Sachdeva N, Weinstein JE, Ashman M, Sachdeva M, Brewer TH, et al. Poor lymphoproliferative responses with low proportion of gag-specific CD8 temra cells in HIV-1-infected patients showing immunological and virological discordance despite prolonged suppression of plasma viremia. Viral immunology. 2010;23(1):49-61.
[Google Scholar] [Cross Ref] [Pubmed]
- Valiathan R, Asthana D. Increase in frequencies of circulating Th-17 cells correlates with microbial translocation, immune activation and exhaustion in HIV-1 infected patients with poor CD4 T-cell reconstitution. Immunobiology. 2016;221(5):670-678.
[Google Scholar] [Cross Ref] [Pubmed]
- Grabmeier-Pfistershammer K, Steinberger P, Rieger A, Leitner J, Kohrgruber N. Identification of PD-1 as a unique marker for failing immune reconstitution in HIV-1-infected patients on treatment. J Acquir Immune Defic Syndr. 2011;56(2):118-24.
[Google Scholar] [Cross Ref] [Pubmed]
- Cuzin L, Trabelsi S, Delobel P, Barbuat C, Reynes J, Allavena C, et al. Maraviroc intensification of stable antiviral therapy in HIV-1-infected patients with poor immune restoration: MARIMUNO-ANRS 145 study. J Acquir Immune Defic Syndr. 2012;61(5):557-64.
[Google Scholar] [Cross Ref] [Pubmed]
- Menkova-Garnier I, Hocini H, Foucat E, Tisserand P, Bourdery L, Delaugerre C, et al. P2X7 receptor inhibition improves CD34 T-cell differentiation in HIV-infected immunological nonresponders on c-ART. PLoS pathogens. 2016;12(4):e1005571.
[Google Scholar] [Cross Ref] [Pubmed]
- Pérez-Santiago J, Ouchi D, Urrea V, Carrillo J, Cabrera C, Villà-Freixa J, et al. Antiretroviral therapy suppressed participants with low CD4+ T-cell counts segregate according to opposite immunological phenotypes. AIDS. 2016;30(15):2275-2287.
[Cross Ref] [Google Scholar] [Pubmed]
- Helleberg M, Kronborg G, Larsen CS, Pedersen G, Pedersen C, Obel N, et al. Poor CD4 response despite viral suppression is associated with increased non-AIDS-related mortality among HIV patients and their parents. AIDS. 2013;27(6):1021-1026.
[Cross Ref] [Google Scholar] [Pubmed]
- Choi JY, Zhou JL, Giles M, Broom J, Templeton DJ, Law MG et al. Predictors and outcomes of HIV-infected antiretroviral-naive patients with discordant responses to combination antiretroviral treatment in Asian and Australian populations: results from APHOD. J Acquir Immune Defic Syndr. 2011;57(1):e13-e15.
[Cross Ref] [Google Scholar] [Pubmed]
- Cen K, Fu LC, Tan XH, Zhen HP, et al. Efect of aiqueqing granule combined with HART on CD4 courts of HIV/AIDS patients with integration integration. Chinese J AIDS STD. 2017; 23(8): 688-690.
[Cross Ref] [Google Scholar]
- Tan Q, Zhou ZH, Yan DM. Immune reconstitution in adult HIV/AIDS patients after long-term antiretroviral therapy. Chinese General Practice. 2020;23(23):2918-2922. [Cross Ref]
[Google Scholar] [Pubmed]
- Lederman MM, Calabrese L, Funderburg NT, Clagett B, Medvik K, Bonilla H, et al. Immunologic failure despite suppressive antiretroviral therapy is related to activation and turnover of memory CD4 cells. J Infect Dis. 2011;204(8):1217-1226.
[Cross Ref] [Google Scholar]
- Ji H, Wang B, Li Q. Fundamentals of management. Posts Telecommun. 2019;79:05.
[Google Scholar]
Author Info
Yong Shuai1,2*,
Hemeng Peng1,4,
Xiaodong Wang1,3 and
Xiaoqing Peng1,3
1Chongqing CEPREI Industrial Technology Research Institute Co., Ltd, Chongqing, 401332, China
2Chongqing Public Health Medical Center, Chongqing, 400036, China
3Chongqing Key Laboratory of Reliability Technologies for Smart Electronics, Chongqing, 401332, China
4Chongqing Institute of Green and Intelligent Technology, Chinese Academy of Sciences, Chongqing 400714, China
Citation: Shuai Y, Peng H, Wang X, Pend X (2022) A Method for the Definition of Immunological Non-Response to Antiretroviral Therapy Based on
Review Analysis and Supervised Classification Model. J Antivir Antiretrovir. S24: 005.
Received: 08-Mar-2022, Manuscript No. JAA-22-16179;
Editor assigned: 11-Mar-2022, Pre QC No. JAA-22-16179 (PQ);
Reviewed: 25-Mar-2022, QC No. JAA-22-16179;
Revised: 28-Mar-2022, Manuscript No. JAA-22-16179 (R);
Published:
04-Apr-2022
, DOI: 10.35248/1948-5964-22.14.005
Copyright: © 2022 Shuai Y, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits
unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.




