Andrology-Open Access
Open Access
ISSN: 2167-0250
ISSN: 2167-0250
Research Article - (2025)Volume 14, Issue 5
Several molecular biomarkers have been proposed to improve prediction of risk on metastasis or on relapse after treatment for cancer patients. However, tumor heterogeneity and high costs of sequencing have obviated the clinical implementation of these molecular markers. In addition, current biomarkers are derived from bulk profiles, which contain both tumor cells and cells from the tumor microenvironment. On the one hand, this results in a confounded biomarker signal influenced by the cellular composition of the sampled tissue. On the other hand, a bulk-derived biomarker does not consider the spatial organization of cells, which has shown important prognostic and predictive potential. Resolving these shortcomings would require expensive spatial profiling, which is infeasible in clinical settings.
Spatially resolved digital profiling, obtained with deep learning models from whole slide images, presents promising potential for cost-efficient exploration of biomarkers at high resolution. Also, the predicted biomarkers from these models are ideal candidates for downstream lightweight, interpretable and efficient clinical outcome prediction. Here, we highlight important guidelines for developing such WSI proxy models, in terms of dataset size and label resolution trade-off, as well as inherent limitations of predicting molecular features on WSIs. We show the added value of molecular WSI proxy models for clinical outcome prediction as opposed to training WSI models directly for outcome, in terms of interpretability, dataset size and model efficiency.
Digital pathology; Deep learning; Digital molecular profiling; Prostate cancer
In prostate cancer, the risk on metastasis after diagnosis or of relapse after an initial treatment remains hard to predict based on clinical features only. Molecular markers, derived by sequencing primary tumor material are increasingly being proposed to guide informed clinical decisions at the time of diagnosis. To avoid missing rare aberrations and to cope with tumor heterogeneity, multiple lesions in the same primary tumor rather than only the dominant lesion should be profiled, but this is cost prohibitive in a clinical setting [1].
In addition, applying sequence-based markers to a tissue results in a bulk-derived signature that captures a confounded contribution of tumor cells and the Tumor Microenvironment (TME). The prognostic or predictive signal of the marker is, therefore, largely affected by the cellular composition of the tissue that was sampled for biomarker profiling. Furthermore, such a bulk signature does not consider the spatial organization of the tumor niche, which is expected to be highly prognostic or predictive. Modern spatial transcriptomics techniques can offer such information, but are excessively expensive for clinical settings [2].
Digital pathology offers great potential as cost-efficient alternative for spatially resolved molecular profiling. Digitized histopathology slides (or Whole Slide Images, WSIs) of the primary tumor are routinely available in clinical care. Studies using deep learning showed that morphological features computationally extracted from these WSIs associate with molecular properties. Consequently, WSIs, in conjunction with WSI-based deep learning models, can predict a tumor’s molecular status in a spatially resolved way without the need for sequencing. These molecular proxies, rather than the true molecular labels, can subsequently be used to predict a disease status.
Here we want to put forward the advantages of WSI-based digital profiling to identify and screen for prognostic or predictive markers and contrast their properties with WSI models that are directly trained to predict clinical endpoints [3].
Impact of model architecture and label resolution on the data requirements of WSI-based models
The development of deep learning models capable of accurately predicting a particular label from WSIs requires either a huge training set of slide/patient-level labeled WSIs (ten) thousands of WSIs) or a smaller training set of labeled WSIs with a clear annotation of the regions from which the label was derived (hundreds of WSIs). These requirements for dataset size and annotation resolution stem from inherent properties of deep learning model architectures for WSIs [4].
Prior to providing a WSI to a deep learning model, the image needs to be subdivided in small patches (tiles) for efficient and effective feature extraction. Tiles are usually taken at 256 × 256 or 512 × 512 pixels at 0.5 μm/pixel, resulting in thousands of tiles per WSI. Depending on the available annotation precision and dataset size, either a tile level or slide level model architecture can be used to predict the molecular label (Figure 1) [5].
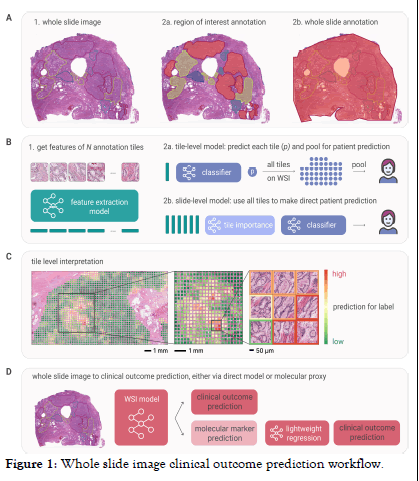
Figure 1: Whole slide image clinical outcome prediction workflow.
Note: Panel A: For a particular whole slide image, labels might be available at some spatial resolution (2a) (e.g., lesion on the image selected for molecular profiling). In some cases (2b), no information is available on which region in the image corresponds to the label of interest (e.g., patient lymph node status). Panel B: To develop a WSI prediction model, first N tiles are extracted from the annotated region(s). Then, a feature extraction model (e.g., ResNet, Vision Transforme) is used to extract features for each tile. In the next step, either a tile-level (2a) or slide-level (2b) model is trained to make a prediction for the label of interest on patient level. The tile-level model requires labels for each tile for training, while the slidelevel model is trained using one single slide-level label. Panel C: Visualizing tile-level predictions for the tile-level model provides a spatial heatmap indicating regions with high prediction values. For slide-level models, techniques exist to approximate/infer tilelevel predictions (e.g., visualizing tile importance). Panel D: To predict a given clinical outcome, two options exist. On the one hand, a dedicated WSI model can be trained directly to predict the desired outcome. Alternatively, the WSI model can be trained to predict molecular markers (e.g., mutations of interest, genetic signatures), which afterwards are propagated to a lightweight, interpretable regression model to predict the clinical outcome.
Tile-level models are trained to make a prediction for each individual tile of the WSI independently. For model training, they therefore require tile-level labels (specific label for each single tile used for training). Most often labels are available at the slide-level only e.g., when considering a patient-level clinical label or a molecular label derived from bulk sequencing a tumor region that was not annotated on the WSI. The requirement of tile level labels therefore necessitates assigning this slide-level label to all tiles in the slide, resulting in a large, labeled dataset of num_tiles × num_slides labeled datapoints that can be used for training (easily >10 k-100 k even for small num_slides ~100). Tile level models can, therefore, be trained on a small dataset of 100s of tile-level labeled WSIs.
However, in case of high tumor heterogeneity, not all tiles in the slide carry the same morphological/molecular properties. Extrapolating a slide level label to each single tile then leads to mis-labeling of potentially thousands of tiles. So, despite the potentially large set of tiles that can be used for training, the noisy labeling prohibits model convergence. It was, indeed, shown that for a heterogenous tumor like prostate cancer, restricting the training to tiles for which the label is more certain significantly increases model performance. Similarly, tile level models that predict molecular labels can directly benefit from fine-grained tile level annotations that are becoming available through modern spatial molecular profiling techniques [6].
If fine-grained annotations are not available or feasible, slidelevel models can be used instead. Such models are trained to make a prediction for each WSI (not tile) and hence require slide-level labels only. These models can derive the relevant tiles for a prediction from the slide automatically, eliminating the need for precise tile-level annotations. However, since these models are trained at slide-level, the number of available training labels is much smaller in this case compared to tile-level models (for a dataset of num_slides WSIs, slide-level models receive num slides labels, compared to num_slides × num_tiles labels for a tile-level model). Reaching robust and accurate model convergence with slide-level models, therefore requires a significantly larger dataset of (ten) thousands of labeled WSIs [7].
For both tile and slide-level models, techniques exist to visualize the predicted label at tile-level. In the tile level model this is achieved by superimposing the tile level predictions back onto the WSI. For slide level models, techniques exist to approximate or infer tile level predictions (e.g., through visualizing the tile importance.
In conclusion, to build robust and accurate WSI prediction models, tile level models can be used in combination with a relatively small set of WSIs, provided the labeling is available at sufficiently high resolution. Slide level models can be used to compensate for the lack of a higher label resolution, but come at the expense of requiring many more labeled WSIs [8].
Predicting clinical outcome with WSI-based models
The aforementioned observations have consequences for the development of deep learning models that directly predict clinical endpoints from WSIs. Since it is unknown up front which histopathological features of the heterogeneous tumor section (region in the WSI) contribute to a patient level clinical endpoint, generating accurate tile level labels is infeasible. Such studies must therefore resort to slide level modeling, requiring paired WSI clinical outcome data of (ten) thousands of patients and an independent resource heavy model training for each new clinical endpoint that is envisaged.
Direct outcome models are therefore only suited for clinical endpoints with routine follow up in standard of care e.g., cancer diagnosis and/or gleason grading, overall survival, metastasis free survival, biochemical recurrence, lymph node status or treatments used in standard of care [9].
They are not applicable for predicting outcome in relatively small sized clinical trials (as not enough training data will be available). Furthermore, direct clinical outcome models are limited in their interpretability. Although feature importance methods can be applied to indicate certain distinctive areas in the WSI which were most decisive for the prediction, these models do not offer straight forward insights into how their prediction corresponds with established biological knowledge relating cellular processes with disease aggressiveness [10].
Rather than directly predicting clinical outcome, WSI-based models can be trained to first proxy molecular labels. Digital profiling of WSIs with these models results in a proxy of the molecular label, which subsequently can be used to predict clinical outcome. Training such WSI-based models for digital molecular profiling requires paired WSI-molecular labels. As the required molecular labels are derived from sequence-based profiling, they have intrinsically a more fine-grained resolution than a patient level label, such as disease outcome. This allows using model architectures (e.g. tile level models) that require a significantly smaller number of labeled WSIs than required for WSI-based models that directly predict clinical outcome [11].
For example, mutational status has been shown predictable with tile-level models from datasets of size ~500 when considering only tiles from the WSI that originate from tumor regions. Performance can be further significantly boosted by further reducing the training to the specific region that was used for sequencing. Soon, development of these WSI-based molecular proxies will benefit from the increasing body of publicly available high resolution molecular labels provided by spatial omics technologies [12].
The molecular markers predicted by these WSI-based models can then be associated to clinical outcome by lightweight and interpretable machine learning models (e.g., logistic regression) (Figure 1d). Such two-step approach for predicting clinical outcome mitigates the mentioned drawbacks of direct outcome models. By using a much simpler model than a WSI deep learning model to perform the eventual association of the molecular proxy with the disease phenotype, significantly less training data is required. This allows predicting disease outcome for smaller sized studies. In addition, the molecular WSI models can be re-used for various clinical endpoints, resulting in higher resource and data efficiency. Finally, they provide a higher interpretability by design, since the prediction for a certain clinical endpoint can be traced back to predicted molecular markers.
WSI-based models trained on mutational signatures capture relevant clinical signals, but underperform as molecular proxies
Indirectly predicting outcome through a molecular label thus depends on WSI-based models trained to predict molecular labels (genetic mutations, gene expression, methylation profiles) from WSIs of tumor samples. Such models predict the probability with which the molecular label is present in each patch of an analyzed WSI. Models have been developed to predict actionable genomic aberrations, with a focus on genes that are frequently aberrant in a pan cancer setting to guarantee sufficient labeled data for training, including somatically mutated genes such as BRAF, TP53 and amplifications in e.g., EGFR. Specifically, for prostate cancer WSI-based proxies for ERG fusions, SPOP and TP53 have been reported. Overall performance in correctly predicting the presence of those aberrations remains modest, also for the more frequently mutated genes [13].
In a case study on prostate cancer (where TP53 is a marker of aggressive disease), it was shown that despite modest performance to predict the TP53 mutation itself (AUC ± 0.7), this predicted mutational status was more significantly associated with lymph node status as a proxy of aggressive disease than the original mutational status determined by sequencing. In-depth analysis showed that the models capture a downstream histopathological phenotype reminiscent of aggressive disease that is characteristic for lesions containing TP53 mutations, but that can also be triggered by other molecular defects (e.g., TP53 deletions or other, more rare alterations).
By observing the aggressive downstream pathogenic phenotype, irrespective of the exact mutation that triggered the phenotype, WSI-based models are seemingly in a better position of predicting disease outcome than when using the original mutational status as biomarker. Their low overall performance as mutational proxy (AUC of in case of TP53) does therefore not interfere with their potential as digital prognostic markers, at least not in prostate cancer. However, this capturing of a downstream phenotype inherently reduces their performance of using histopathological features for proxying the mutational status itself [14].
WSI-based models trained on expression signatures represent reliable molecular proxies
Given these observations made for WSI-based proxies of TP53 mutation in prostate cancer, we argue that when trained on molecular labels that better reflect downstream pathways, such as expression labels, WSI-based models have potentially the same prognostic or predictive value, while at the same time being good molecular proxies. Several endeavors have already been made with models that proxy expression signatures, either by using a model architecture that learn from all genes together or models that are trained to predict the expression of each gene or a subset of genes only. The genes for which good WSI-based proxies of gene expression can be obtained differ per cancer type and in general mark cell types with visible features on the WSI, such as endothelial cells and immune cells or genes involved cancer hallmarks.
Good WSI-based molecular proxies allow coping with tumor heterogeneity and with the limited statistical power that so far obviated the clinical implementation of many previously described molecular markers. Further, it opens avenues for the cost efficient screening and validation of previously described molecular markers in large patient cohorts or for the validation and/or detection of novel biomarkers in clinical trials for which no sequencing data is available.
Furthermore, they enable for the first time to investigate the prognostic and predictive value contained in the spatial colocation of molecular markers and their associated cell types. A recent study shows how mapping the tile level predicted gene expression made by the model on the original WSI provides a proxy of spatially resolved transcript profiling that approximates well the expression patterns observed by true spatial transcript profiling. In addition, the expressions of cell type marker genes are predicted to be spatially collocated on the WSI, indicating that the deep learning model can extract the co-expression relation between these genes by their association to similar features on the WSI [15,16].
Future improvements
Here, we presented the advantages of using molecular WSI models as proxy for clinical outcome prediction as opposed to directly training models on WSIs for clinical outcome. While currently released molecular WSI proxy models show significant potential, further improvements are necessary to enhance the accuracy and robustness of the results.
Here, we presented the advantages of using molecular WSI models as proxy for clinical outcome prediction as opposed to directly training models on WSIs for clinical outcome. While currently released molecular WSI proxy models show significant potential, further improvements are necessary to enhance the accuracy and robustness of the results [17].
Particularly, molecular WSI proxy models can benefit greatly from the growing availability of fine-grained labels obtained with modern spatial transcriptomics. Spatial transcriptomics data provides ground truth at tile level, which can directly be used to fine tune tile level models. Also, these labels can be used to indirectly guide the tile importance mechanism of slide-level models. We expect that such incorporation of spatial transcriptomics labels into existing pipelines will boost performance significantly [18].
Apart from increased label resolution, various improvements in model architecture can also aid in increasing robustness of results. Currently, the most widely used tile feature extractors in WSI models are ResNet variations pre-trained on ImageNet (14 million hand-annotated images from over 20.000 categories ‘cat’, ‘dog’...). However, image features derived from ImageNet are very different from those in WSIs, where morphological features at different scales occur. Because of the different nature of features, the use of (partly) fixed feature extractors from ImageNet might lead to missing relevant morphological features in WSIs. Recently, several pre-trained feature extractors have been proposed which have been pre-trained on several hundred thousand patches in a self-supervised learning setting [19,20]. We expect these specialized WSI feature extractors to play an important role in improving accuracy and robustness of the predictions. Further, improvements in training schemes and more efficient architectures are active research areas which have consistently improved performance in WSI prediction tasks.
In conclusion, we expect that through both novel model designs and increased label quality, the accuracy and robustness of existing models for digital spatial expression profiling from WSIs will improve significantly. This opens avenues for not only cost efficient analysis of tumor heterogeneity and exploration of gene expression dynamics at high resolution, but also for downstream lightweight, interpretable and efficient clinical outcome prediction.
M.P. was supported by a grant from FWO 1161223N and FWO V467423N. The work was further supported by grants of the Fonds Wetenschappelijk Onderzoek-Vlaanderen (FWO) (3G045620, 3G046318), SBO (S004824N) and UGent BOF (BOF 01J06219, BOF/IOP/2022/045BOF).
The a uthors declare no competing financial or non-financial interests.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Pizurica M, Marchal K (2025) The Future of Digital Biomarker Profiling. Andrology. 14:355.
Received: 16-Apr-2024, Manuscript No. ANO-24-30807; Editor assigned: 19-Apr-2024, Pre QC No. ANO-24-30807 (PQ); Reviewed: 03-May-2024, QC No. ANO-24-30807; Revised: 02-May-2025, Manuscript No. ANO-24-30807 (R); Published: 09-May-2025 , DOI: 10.35248/2167-0250.25.14.355
Copyright: © 2025 Pizurica M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.