Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Review Article - (2023)Volume 16, Issue 3
In silico vaccine design is the computational biochemical preparation of vaccines. It is a modern approach to vaccine design and is very fast, inexpensive, effective and applicable to antigenically diverse pathogens when compared to the traditional method of vaccine design. The aim of this review is to explain the important steps, bioinformatics tools and immune-informatics approach useful in computational vaccine design. The databases and algorithms include National Centre for Biotechnology Information (NCBI), EMBOSS transeq, ANTIGENpro, AllerTOP, Toxinpred, NetCTL 1.2 server, IEDB MHC-II, and BCPred, which are respectively used for sequence retrieval and analysis, sequence translation, antigenicty, allergenicity and toxicity prediction, Cytotoxic T-Lymphocyte (CTL), Helper T-Lymphocyte (HTL) and B cell epitopes prediction. Additionally, this review gives a detailed report on the processes of the construction of vaccine primary and secondary structures, vaccine physico-chemical properties, tertiary structure prediction, refinement and validation, stability enhancement, molecular docking and dynamic simulation, immune response simulation, codon optimization and cloning. In conclusion, this report allows for a comprehensive insight on subunit vaccine construct.
In silico; Vaccine constructs; Computational tools; Databases; Bioinformatics; Traditional
Vaccines are biological preparations that have saved millions of lives by training our immune systems to fight infectious pathogens [1]. Vaccine design is broadly categorized into traditional approach and modern approach. The traditional vaccine design is costly, ineffective, time-consuming and inapplicable to pathogens of diverse antigen [2]. On the other hand, the modern vaccine design is relatively inexpensive, fast, effective and applicable to antigenically diverse pathogens. This is made possible through advanced modern technologies that have facilitated computational or in silico method of vaccine development, including recombinant Deoxyribonucleic Acid (DNA) technology, rational vaccinology, structural biology, conjugate vaccines, Next Generation Sequencing (NGS) technology and epitope-based vaccine design, which applies for the bulk production of sub-unit vaccines [3].
Bioinformatics is a field that manages, analyzes, interprets, and uses biological data through information technology and mathematical concepts. It is a field that is continually developing and creating helpful tools for biological sciences [4]. Critical study of biological data is important for understanding the metabolic, pathological, physiological, and anatomical and biochemical activities of living organisms [5].
Bioinformatics tools are computational algorithms and software tools essentially utilized to understand, describe, predict or manipulate biological processes, with the primary goal of serving the agriculture and pharmaceutical industries, health care, forensic analysis, crop improvement, food analysis, vaccine design, drug discovery, and biodiversity management [6].
This review gives general steps, algorithms and immununo-informatics approach used for primary, secondary and tertiary construct, codon optimization and in silico cloning of a subunit vaccine.
The following are series of steps and computational tools necessary for vaccine construct.
Retrieval and analysis of sequences
There are enormous public online databases where researchers can readily access molecular sequences; this is the essential first step in in silico vaccine design, after determining the disease of interest and the antigenic part of the pathogen. The available public DNA and protein sequence databases are huge, hence, the importance of sequence retrieval and analysis. After identifying the antigenic part of the disease of interest (pathogen), the nucleotide sequences are retrieved, which can be achieved via global nucleotide sequence storage including National Centre for Biotechnology Information (NCBI), European Molecular Biology Laboratory (EMBL) and DNA Database of Japan (DDBJ) databases.
These databases contain essentially the same data; others include protein sequence databases like UniprotKB, Swiss-Prot, TrEMBL and Protein Data Bank (PDB). Some pathogens have specialized database for sequence retrieval, for example, influenza virus, COVID-19 and Respiratory Syncytial Virus (RSV) amino acid sequences are reliably retrieved from Global Initiative on Sharing All Influenza Data (GISAID) [7]. The sequences should be downloaded and saved as FASTA format and its accession number should be noted [8].
Annotation of retrieved sequences: Sequences annotation is important in cases where the whole genome sequences of the pathogen was retrieved, this step will assist to identify previously established antigenic part of the pathogen by aligning your retrieved sequences with a reference sequence through sequence alignment, this process is referred to as sequence analysis.
One of the main study fields in computational biology is computational genome annotation of sequences, which has grown in importance with the introduction of high-throughput sequencing technology. Sequence annotation phase might not be necessary if you only retrieve the antigenic part of the pathogen; however, if you retrieve the pathogen’s complete genome sequences, sequence annotation is crucial to determine the genes functions and the genome's evolutionary history [9].
When DNA, Ribose Nucleic Acid (RNA), or protein sequences are arranged in order to detect regions of similarity, which may be the result of functional, structural, or evolutionary links between the sequences, this process is known as sequence alignment. Finding similarities between different sequences is a key objective in genomics. In order to determine the function of a sequenced gene, sequence alignment can be used to compare the sequence to similar sequences from a database. Sequence alignment gives information on the level of similarity between two sequences and the approximate length of time within which the two sequences diverged [10,11]. Additionally, sequence alignment is also important as a common first step in other processes like phylogeny building.
For vaccine construct, pairwise sequence alignment tools are employed in sequence annotation which includes; Smith-Waterman algorithm (SSEARCH), Fast All (FASTA), Basic Local Alignment Search Tool (BLAST), EMBOSS tools and Dot plots [12]. Bioinformatics tool for multiple sequence alignment includes CLUSTALW, Constraint-based Alignment Tool (COBALT), Multiple Sequence Comparison by Log Expectation (MUSCLE), ProbCons, Multiple Alignment using Fast Fourier Transform (MAFFT) and T-Coffee.
Translation of nucleotide sequence to protein sequence: In the process of vaccine construct, it is important to translate nucleotide sequences to protein sequences and tools used are European Molecular Biology Open Software Suite (EMBOSS) transeq, Expasy, DNA to protein translation, Vector Builder used for various biological analysis including sequence alignment [13].
Prediction of antigenicity, allergenicity and toxicity
Antigenicity screening helps us to predict whether the protein sequence is strong enough to be recognized as a non-self in the human system and thus stimulate cellular and humoral immune response [14]. The major servers employed for this function are ANTIGENpro on the scratch protein and VaxiJen v2.0 [15]. Sequences meeting the threshold for antigenicity (≥ 0.8 on ANTIGENpro and ≥ 0.4 on VaxiJen) are considered antigenic, and qualified to be used for further analysis [16].
Allergenicity screening helps to determine the allergenic behavior of sequences. In order to be qualified for further analysis, sequences must be non-allergenic in nature. Software servers for this screening are AllergenFP and AllerTOP, overall accuracy based on the statistical analysis of sensitivity and specificity confirms that they are the best allergen prediction tools [17]. The sequences must be not being toxic to the human system; the prediction server for toxicity is ToxinPred. ToxinPred utilizes dataset of 1805 toxic peptides from 35 or less residues to test the toxicity of query peptides [18].
T and B cells epitopes prediction
Epitope determines the antibody (T and B lymphocytes) attachment site on the part of antigen and is recognized through the host immune cells. The essence of vaccine is to be able to prompt cellular and humoral immunity, hence, the necessity for T and B cells epitopes prediction. A term used for mapping of B cell and T cell epitopes, Epitope fishing can screen the potential epitope in a pathogen [19]. Mapping epitopes that could be incorporated into the vaccine is based on several formulas and qualities.
There are two types of T cell, Cytotoxic T cells (Tc cells) and Helper T cells (Th cells) and the pathways of preparing and introducing epitopes to the two sorts of T cells are unique [20].
Cytotoxic T Lymphocyte (CTL) epitopes prediction: NetCTL 1.2 server is used to locate CTL epitopes [21]. The prediction efficiency of the server is determined by the proteasomal C-terminal cleavage, Transport associated with Antigen Processing (TAP) efficiency, and Major Histocompatibility Class I (MHC-I) affinity parameters. Default value for weights on C-terminal cleavage is 0.15, TAP transport efficiency is 0.05, and default threshold value for epitopes is 0.75. TAP transport efficiency uses weight matrix while peptides binding to MHC-1 and proteasomal C-terminus cleavage are achieved by Artificial Neural Networks (ANN). When making use of default threshold, epitopes with ≥ 0.75 are selected for further analysis.
Helper T Lymphocyte (HTL) epitopes prediction: Immune Epitope Database (IEDB) MHC-II is used to predict HTL epitopes [22]. A reference set of alleles in form of human MHC alleles, including Human Leucocyte Antigen (HLA) A, B, C and E, or animal alleles (Monkey, Cattle, Pig, Mouse) is required at this stage to help select HTL epitopes [23].
The epitope sequence with the lowest percentile rank for each allele used, as depicted by IC50, usually shows higher binding affinities towards the allele’s HTL receptor, so they are selected for incorporation into the vaccine construct.
B lymphocyte epitopes prediction: The B cell epitopes prediction is done via online tool server, BCpred, all parameters including the length and specificity of each epitope can be set or left at default, the peptides, scores, and start position are shown by the predicted B cells [24]. Non-overlapping epitopes with highest scores are selected. Other tools used in B cell epitopes prediction are ABCPred, IEBD, BepiPred, LBtope, DiscoTope, Ellipro, Epipred.
Overlapping epitopes selection, antigenicity, allergenicity and toxicity screening for CTL, HTL and B cells epitopes: According to Oluwagbemi, et al. [12] research, when the epitopes gotten for CTL, HTL and B cell were numerous, overlapping occurred, therefore, epitopes with a frequency of overlap occurring 60 times and above was screened down, and these epitopes were selected for the vaccine construct. The predicted CTL, HTL and B cell epitopes are subjected to antigenicity, allergenicity and toxicity screening, while the epitopes that are antigenic, non-allergenic and non-toxic are selected for further analysis.
Construction of vaccine primary structure
CTL, HTL and B cell epitopes are compiled into subunit vaccine; each is linked with a brief amino acid called Linkers. In order to enhance the immunogenic potential of the vaccine, an adjuvant is placed at the begininning (N-terminus) or end (C-terminus) of the vaccine peptide sequence, or both sides [25]. The widely used Linkers are KK, AAY, GPGPG and EAAAK, though their purposes vary. These linkers are crucial in decoding the site which confers expression of epitopes and enhance immune responses to vaccine [26].
AAY, GPGPG and KK linkers are used to join CTL, HT, and B cell epitopes, respectively. EAAAK is a rigid linker used for maintaining distance between domains to improve protein stability. EAAAK sequence is placed immediately after the adjuvant sequence and used to separate epitopes from the adjuvant; it may also be placed at both end of vaccine peptide sequence. CTL epitopes are placed before HTL epitopes; note however that B cell epitopes can be placed near N or C-terminus but not in between the T cell epitopes. Adjuvants are selected in relative to the pathogen studied and the desired immune response, commonly used adjuvants are RS09, Saponin, 50S L7/L12 ribosomal protein, Hp91, Cholera toxin subunit B (ctxB), Heparin-binding hemagglutinin and many others [27-32]. Adjuvant+Linkers+Epitopes organization is the ultimate vaccine design (Figure 1).
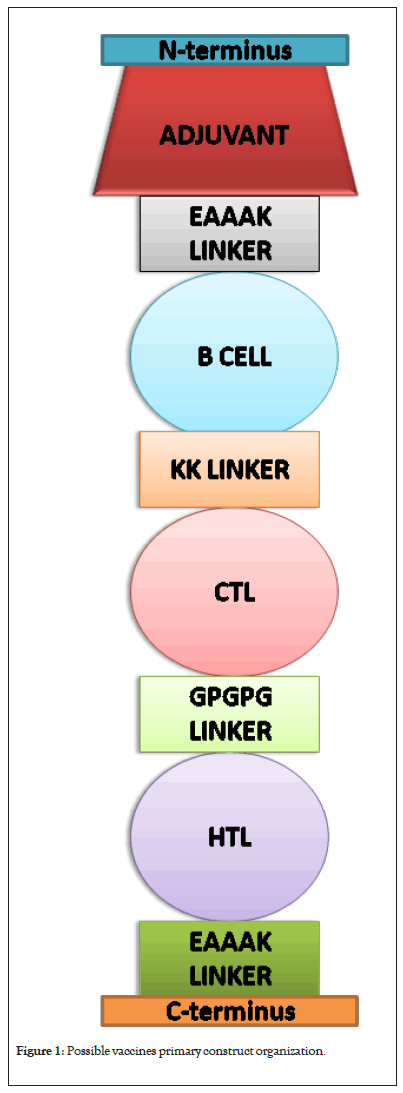
Figure 1: Possible vaccines primary construct organization.
Screen the constructed vaccine primary structure for antigenicity, allergenicity and toxicity effect, a well-designed vaccine is generally regarded as safe when it is non-toxic, antigenic yet anti-antigen, anti-allergen/non-reactogenic, and capable of accelerating immunity [33].
Physicochemical properties of vaccine construct
This is done to elucidate the physical and chemical nature of the vaccine in terms of its molecular weight, aliphatic index, Instability Index (II), Grand Average of the Hydropathicity (GRAVY) value, theoretical Protrusion Index (PI), solubility, and estimated half-life [34]. ProtParam webserver is used to determine these properties (in vitro and in vivo), it is an Expasy suite that consists of bioinformatics tools [35]. Also, solubility of the vaccine can be cross-checked by using SOLpro, an online tool available in the SCRATCH suite [36].
Construction of vaccine secondary structure
Vaccine primary construct is subjected to specify online server tools to predict the vaccine secondary structure, the tools include; Self-Optimized Prediction Method (SOPMA), the conformational states assessed by SOPMA were the helices, sheet, turn and coil of the construct, also, PSIPRED server estimates the secondary structure of the vaccine sequence [37,38].
Tertiary structure/3D structure prediction, refinement, validation and flexibility analysis
The vaccine primary construct is subjected to specified online server tools to predict the vaccine tertiary structure; it is regarded as vaccine 3D structure. The prediction is essential in order to better understand the vaccine biological functions. Iterative Threading ASSEmbly Refinement (I-TASSER) is used to able and generate 3D models of proteins on the basis of sequence to structure to function paradigm and has been ranked as one of the best servers for protein structure prediction [39]. There are several vaccine 3D structure prediction including SWISS-MODEL, Phyre2 and RaptorX [40].
3D structure refinement is important to enhance the structural and global quality of the 3D structure by reconstruction, repacking and simulations of the 3D structure for relaxation. ModRefiner and GalaxyRefine tool of the GalaxyWeb server are used for initial and secondary refinement in order to get a more refined structure. It should be noted that the selected refined 3D structure should be downloaded and saved as PDB format [41,42].
In order to validate the refinements done, servers are employed to perform the Ramachandran plot analysis, Z-score analysis and assessing potential errors in the 3D crude model. ProSA-web, RAMPAGE and ERRAT webser can be employed in the validation of the refined 3D structure. RAMPAGE server probes Ramachandran plot and the principles of PROCHECK for validation through Ramachandran plot. At the same time, Prosa-Web and ERRAT highlights Z-score analysis and excellent scores of the calculated errors in the queried 3D structure [43].
Furthermore, the flexibility of the vaccine is analyzed by an online tool CABS-flex 2.0, this tool perform 50 rounds of simulation at a default temperature of 1.4°C, It effectively perform simulations of a protein’s structural flexibility [44].
Prediction of linear and conformational B-cell 3D structure
The projected tertiary structure is subjected to Ellipro to determine the B-cell 3D linear and conformational structure. This tool uses algorithms to determine PI of residue, to estimate protein shape and cluster formation of the neighboring residues. Each predicted epitope is designated with a PI value that was averaged over the epitope residue and discontinuous epitopes on the protein structure is visualized through an open-source molecular viewer Jmol [45].
Stability enhancement
Stability enhancement is aimed at improving the stability and strength of the refined tertiary protein structure, the process used to achieve this is known as disulfide engineering. Input the refined protein structure in Disulfide by Design 2 server (DbD2) available online, set all parameters on the server todefault, and select the best mutants possible on energy level and Chi³ value [46,47].
Molecular docking
Molecular docking is a computer simulation used to predict the binding ability of two molecules. It could be protein-ligand docking as in drug design, or protein-protein docking as in vaccine design.
The refined tertiary vaccine structure (protein) is docked against a novel human receptor (protein) to predict the vaccine-receptor binding ability. Most commonly used receptors are Toll-like Receptors (TLR) and MHC class II receptors, these receptors are downloaded from Protein Data Bank (PDB) database and must be cleaned before further use. Server employed for protein-protein docking are PatchDock, ClusPro 2.0 server, and HDOCK [48,49]. Also, after obtaining your result from ClusPro 2.0 server, select the best model on the basis of any criterion (balanced, electrostatic favored, hydrophobic-favored, VdW+Elec) as you desire, these models are viewed with the aid of a molecular viewer (e.g. pymol or jmol viewer). The binding affinities between the vaccines construct and immune receptors can be cross-verified using PatchDock webserver which uses a rigid body algorithm and the top 10 results generated by Patchdock are submitted to FireDock for further improvement [50,51].
Molecular Dynamics (MD) simulation
This is vaccine-receptor simulation, this analysis is done to obtain the biophysical interactions between the vaccine, its surroundings, and the receptors (MHC-II or Toll-like receptors), and the tool used is Groningen Machine for Chemical Simulations (GROMACS) software. Similarly, iMOD server is also utilized for MD simulation to predict the stability of interaction between the receptors and the vaccine construct. Interpretation of the stability of the vaccine-receptor complex is done from output parameters, such as deformability, eigenvalues, and covariance.
Immune response simulation
This stage is important for prediction of the vaccine’s ability to elicit an effective immune response if it is administered in a real-world setting, it indicate if the designed protein could generate positive immune responses and causes considerable spike in levels of antibodies. C-ImmSim online server is an agent-based algorithm for the estimation of antigen and immune activity; it utilizes a Position Specific Scoring Matrix (PSSM) to help analyze the magnitude of immune response generated as a result of vaccine dosage at different time intervals [52,53].
Codon optimization and in silico cloning
For codon optimization, it is important to adapt the vaccine construct to the gene of the microorganism of interest; this allows conformity between them, thereby resulting in potency and high levels of activity. Java Codon Adaptation Tool (JCAT) is used for this purpose, it which corroborates codon compliance by optimizing the vaccine construct sequence. The CAL-Value and GC-content of the improved sequence should be noted, these must be approximate to 1.0% and 50 above. DNA sequence generated, cDNA, should also be noted as this will be used in in silico cloning on SnapGene software application.
In silico cloning is done to carry out expression analysis, the designed vaccine construct is cloned inside the host vector or micro-organism of interest. For cloning, vector modification N and C-terminal of the vaccine are performed with two restriction enzymes. And for expression, the generated optimized DNA sequence (cDNA), along with the restriction sites were incorporated into the pET-28a (+) vector utilizing the Snap-Gene software application tool [54].
Population coverage
Bioinformatics tool for this is IEDB population coverage analysis, found in the weblink http://www.iedb.org/. Using MHC binding or T cell restriction data and is Human Leucocytes Antigen (HLA) gene frequencies , the server calculate the percentage of people in that location who have shown possible responses to the T cell epitopes.
When the alleles used for epitope prediction is human leucocytes antigen, then T cell epitopes selected for vaccine design must be predicted for their ability to binds to HLA molecules across different human population and the likelihood of it inducing long-lasting immune response in this population. IEDB population coverage tool is used in predicting T cells epitopes with high percentage of been presented in the context of HLA molecules to induce immune response in population (Figure 2) [55].
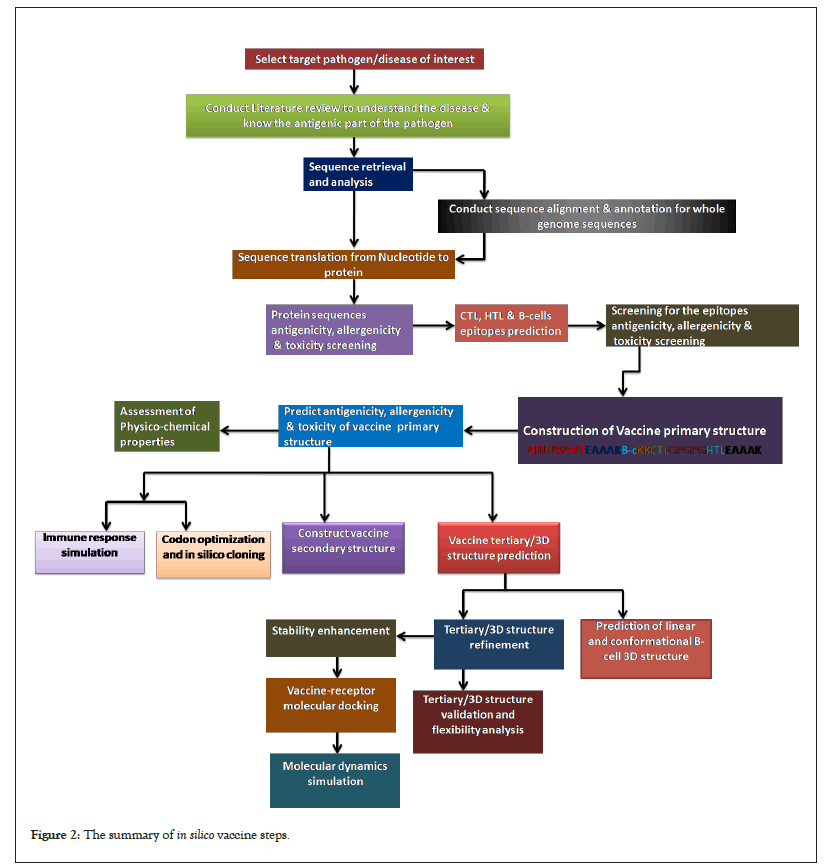
Figure 2: The summary of in silico vaccine steps.
Additional data analytics tool
R programming language is an analytical programming tool that can be utilized in the process of vaccine construct. According to Oluwagbemi, et al. [12] research, in order to obtain the distribution of the clades and lineages for sequences of interest, R Studio was utilized to subject the metadata retrieved from GISAID database to epidemiological analysis.
Vaccine development is a significant aspect of global public health. Traditional method of vaccine design is expensive and not applicable to pathogens of numerous antigens, this had been a major limitation in the past. The advancement in modern technologies is gradually filling this gap through the discovery of computational vaccine design method, which is fast, inexpensive and very efficient in preventing diseases, thus saving millions of lives. Furthermore, application of bioinformatics techniques has increased the availability of pathogenic microbe genome sequences which results in discovery of genes and proteins that could be use in drugs or vaccines development. This report gives a general insight on the databases, software tools and the consequential steps used in retrieving of sequences and locating T and B-cell epitopes that will result in the development of vaccine. Further study and explanation that will elaborate and simplify the several aspects of in silico vaccinology is recommended.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Olawale B, Oyewole TA (2023) Steps and Tools Involved in In Silico Vaccine Design: A Review. J Proteomics Bioinform. 16:647.
Received: 28-Aug-2023, Manuscript No. JPB-23-25784; Editor assigned: 30-Aug-2023, Pre QC No. JPB-23-25784 (PQ); Reviewed: 13-Sep-2023, QC No. JPB-23-25784; Revised: 20-Sep-2023, Manuscript No. JPB-23-25784 (R); Published: 29-Sep-2023 , DOI: 10.35248/0974-276X.23.16.647
Copyright: © 2023 Olawale B, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.