Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research Article - (2021)Volume 14, Issue 3
Data sharing is critical for open science and often required by funding organizations and journals. NCI has developed the Proteomics Data Commons (PDC) as part of the Cancer Research Data Commons, an infrastructure that allows users to share, analyze, and store results, utilizing the storage and compute resources of the cloud. To date most of the data available in the various Data Commons are submitted from large multi-institution research programs funded by NCI with teams of specialists from multiple scientific disciplines. Here we describe our experiences and summarize the recommended best practices for sharing a set of proteomics and related biospecimen data and analyses results from smaller scale proteomics studies conducted in an academic medical center core facility using patient samples of lung adenocarcinoma. Mapping and depositing data in the manner described here harmonizes user’s data to a common data model and community standards, making it possible to view the data alongside other high value cancer studies available in the PDC.
Availability: Data, metadata, protocols with peptide and protein identifications are available at the PDC. (https:// pdc.cancer.gov/pdc/study/PDC000231).
Proteomics data sharing; Best practices; Proteomics resources
Data sharing is critical for open science and increasingly required by funding organizations, journals and the scientific community in general [1,2]. NCI has developed the Cancer Research Data Commons [3] as an infrastructure that provides secure access to many different data types across scientific domains, allowing users to share, analyze, and store results, leveraging the storage and compute resources of the cloud. These resources provide valuable data sets in a Findable, Accessible, Interoperable and Reusable (FAIR) manner, [4] to the global cancer research community in standardized formats and data models. However, most of the data available in the various Data Commons are submitted from large multi-institution research programs like TCGA, [5] CPTAC [6] and others that have teams of specialists in the technologies (i.e. genomics, proteomics, imaging), data management, data science, statistics, clinical science and more who can facilitate the submission and sharing of data. How practical is it for bench scientists at smaller medical research centers to submit and share research data from their local biospecimen and proteomics research core facilities? Here we present a case study to summarize our experience and suggested best practices for submitting a set of proteomics and biospecimen data from lung adenocarcinoma tumor samples from research conducted at Georgetown University’s Lombardi Comprehensive Cancer Center (LCCC).
Twenty samples of frozen lung adenocarcinoma tumor and adjacent normal tissue from 10 individuals and related specimen, demographic and diagnosis metadata were obtained from the Histopathology & Tissue Shared Resource at LCCC after approval from the institutional Biospecimen Use Committee. Quantitative tissue proteomics was performed using modified CPTAC protocols developed for lung adenocarcinoma [7], including Optimal Cutting Temperature compound (OCT) removal, bottom-up proteomic sample processing, and nanoUPLC-MS/MS analysis on a TripleTOF 6600 mass spectrometer (Sciex) coupled with a nanoAcquity UPLC system (Waters) [8]. Three sets of 8-plex iTRAQ-labeled samples were analyzed (with 24 fractions from each set), yielding a total number of 144 raw files. Identification and quantification of proteins were carried out with Protein Pilot 5.0 software [9], by searching against the human proteome sequences from UniProtKB/Swiss-Prot database [10,11]. Details of the methods and all results (including the raw and processed data, initial peptide and protein identifications and deidentified biospecimen data with limited clinical information and written descriptions of the sample and analysis methods) have been submitted to the Proteomics Data Commons (PDC) and the documents are publicly available for download. Details of this submission process and the sharing of all data are described here. Detailed biostatistics/bioinformatics analyses of the results will be published later.
Data mapping
The first step in submitting to the data commons is to map the experimental data, files and associated metadata to the resource’s data model and terminology. Completing this step as comprehensively as possible provides for a smooth submission process and allows interoperability with other data in the data commons ecosystem. The PDC provides its data model and data dictionaries online as well as directions for data submission (https://pdc.cancer.gov/pdc/submit-data) along with training videos. Here we provide our experience and recommendations on how to best navigate and complete the process. Wherever possible, the PDC data dictionaries use community-accepted vocabulary and nomenclatures from the Cancer Data Standards Registry and Repository [12], NCI Thesaurus [13], and the Proteomics Standards Initiative [14] to annotate clinical attributes, peptides, proteins, modifications and Mass Spec related attributes. The PDC also provides a submission workbook with example values and has each column hyper-linked directly to the online data dictionary for details on format and terminology options for mandatory and optional data elements. Completing the submission workbook as completely and accurately as possible is key to a smooth successful submission. We strongly recommend resolving any missing items and uncertainties in terminology before proceeding beyond the initial registration with the PDC. The PDC has curation help available at nci.pdc.help@esacinc.com to answer any questions from users. We recommend consulting with them about questions on formatting or terminology.
It is important to map the sample metadata to the closest terminology in the data dictionary. This can be challenging the first time as terminologies in use are different between institutions, and even local terminologies and sample collection and preparation methods within an institution may change over time and recorded differently depending on when the samples were obtained. Users should be prepared to go back to their biospecimen repository and inquire about some items for clarity. For example, we inquired about freezing_method and method_of_sample_procurement and similar terms. The repository manager had useful answers and suggestions on terminology choices. Note, repository data dictionaries are not perfect and changes can be requested and alternate terms suggested. For example, we did not find the terms “adjacent normal tissue” or “tumor adjacent normal” we expected to capture for our dataset so used “Solid Tumor Normal”; however, after communicating with the PDC, the data dictionary was updated and now includes “adjacent normal tissue”.
One very important step is to ensure that any patient samples are adequately deidentified. The PDC will only accept deidentified data and will screen for HIPAA prohibited data elements [15] but it is the submitters’ responsibility to check these as well and remove or properly anonymize them. Academic and hospital biospecimen repositories typically provide only deidentified data with the samples. However, data submitters should screen the information themselves and remove or change any internal institutional sample or case identifiers because, a) they may encode information, like a date or year sampled, and b) another individual at your institution who has access to patient information could recognize the identifier format and query the patient data. We recommend that users create and submit new unique identifiers that encode nothing about the sample to replace those provided by their biospecimen core. Users may need to create unique file names with their submission as some analysis pipelines reuse names for each run but store them in directories with different names, which can be confusing when transferring or using downloaded files. The submission workbook helps resolve the organization of sample runs and names before the submission. Figure 1 outlines our recommended process for data submission.
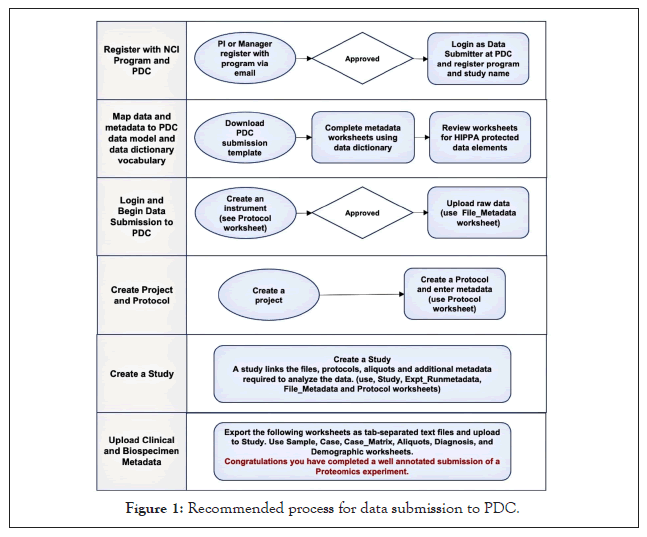
Figure 1: Recommended process for data submission to PDC.
Data Submission: Once the data submission spreadsheet is complete with values for all required data elements, which may be listed as “not reported” in some cases, users can login, start entering data and load files following the directions and videos provided by the PDC. For efficiency, we recommend that the users follow the recommended steps in the order provided in the instructions. First step is to submit information on the mass spectrometry instrument(s) followed by uploading the required instrument specific data files from the instrument either directly from the user’s computer or from Amazon s3 buckets. The PDC has an interactive interface for these tasks and most of the information needed can be copied from user’s data submission spreadsheet. Next, metadata on the project, protocol, and study design need to be provided. Data uploaded into PDC, including the files and metadata, are validated automatically as per the PDC dictionaries and provides clear error messages that users must address to continue the submission. Finally, the user can submit biospecimen metadata including information on cases, demographics, diagnosis, samples and aliquots. For this data the user can export the submission spreadsheets individually to *.tsv format (tab-separated values) and load them directly, no copy-paste required. If user wishes to share more information such as the results of the peptide and protein identification, documents on the sample prep, and detailed documents on methods, users can submit them as additional metadata files or contact the PDC for submission of these files. This completes the submission process for raw data and a ‘complete’ set of experimental and biospecimen metadata. Your submission is initially private. It can be made public immediately on your approval or following acceptance of a publication.
All the raw datafiles, methods, protocols, sample metadata, plus peptide and protein identifications from this study were submitted following the steps described above and are publicly available at the PDC (https://pdc.cancer.gov/pdc/study/PDC000231) and through the PDC APIs for anyone to examine and use in their research. Figure 2 shows some of the data files and metadata files we uploaded to the PDC. The current PDC data model has 215 data elements with approximately 80 required elements. The exact amount varies with instrument and methods with higher multiplexed methods like 16-plex TMT requiring more. For our work, we submitted all 80 elements required plus another 21 we had available or obtained from our institutional databases. Thirteen of the required data elements were filled as “not reported”. This covered 20 samples plus controls and 144 machine files. We also chose to submit additional documents describing our methods and identifications for peptides and proteins. We feel this additional information is best practice that should be encouraged.
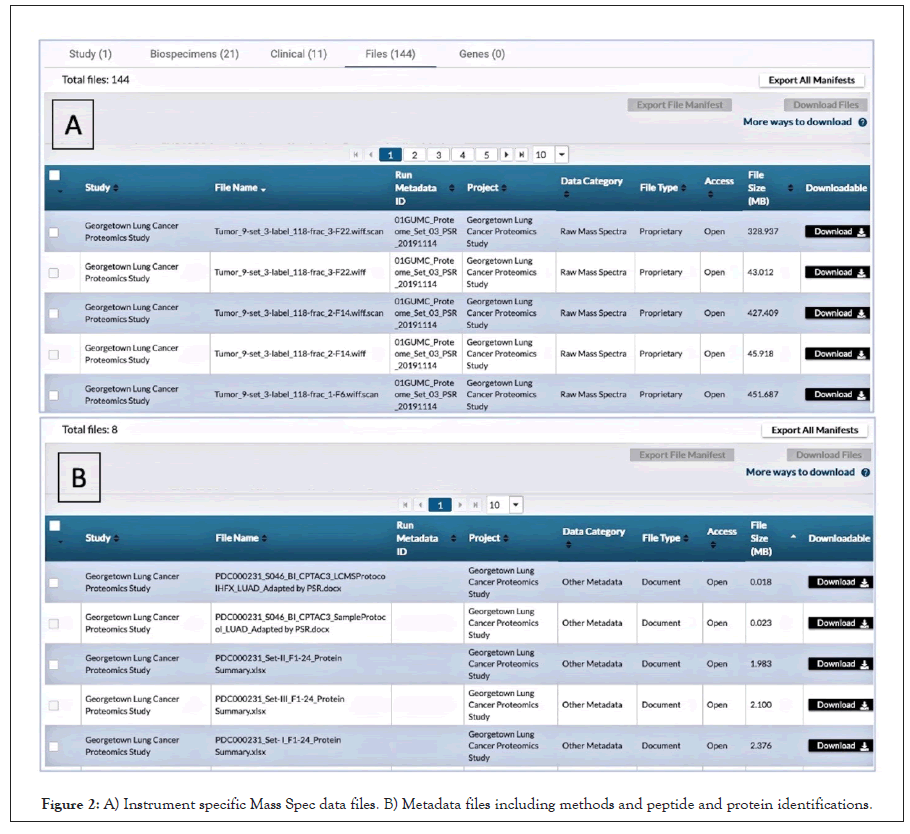
Figure 2: A) Instrument specific Mass Spec data files. B) Metadata files including methods and peptide and protein identifications.
We did observe some minor bugs, usability and documentation issues that we communicated back to the PDC. Many of these have been addressed or are in progress. We found copy/pasting all the file metadata time consuming but that has been improved and additional improvements are under development. The PDC has only become public in the last year and plans to conduct user surveys, workshops and usability studies to make additional improvements as more data is submitted and more users download data through the website and APIs.
The submission process does take some time and effort from one or more individuals, especially the first time. Submitters need to understand the data model and data dictionary options, collect additional data from your proteomics and biospecimen cores, rename files if necessary, check that your data is adequately deidentified, create unique identifiers you may not have for samples, aliquots, cases etc., and reviewing the workbook prior to submission. However, this is time and effort well spent and still is a fraction of the effort of preparing this or any manuscript for publication and arguably as important to reproducible science. Experience with biomedical informatics and proteomics methods helps. Submission of mass spec proteomics data often requires the assembly of information from individuals in different laboratories who may not be directly involved in the overall research study so open communication between labs is essential.
Though there has been great progress on developing community data standards (ontologies, controlled vocabularies and file formats) for data harmonization and FAIR data exchange, this remains a challenge for researchers and repositories as multiple standards are in use, even within institutions. Efforts to harmonize terminologies and automate the process are ongoing. NCI has created a Center for Cancer Data Harmonization [16] that will assist in harmonization of data and terminology available throughout NCI's Cancer Research Data Commons. Currently there is no simple scalable solution to both effectively collect enough metadata to improve data reuse and also reduce the burden of data submission. However, there are approaches that can help: 1) Researchers should identify the target repositories at the start of their research project and try to follow data standards from the beginning of the data life cycle not at the end prior to publication; 2) Adopt community standards, internally at the data collection points such as the biospecimen core repositories, genomic or proteomic analysis cores; and, 3) There are resources to help select terminologies, formats and databases, for example www.FAIRsharing.org [17] and also online database tools such as the NCImetathesaurus (ncimeta.nci.nih. gov/ncimbrowser/) [18] and BioPortal (bioportal.bioontology. org) [19] can search and convert between biomedical terminologies and ontologies. Eventually there will more be automated tools to help with data harmonization and ease the time required for data submitters (Figure 3) [20-23].
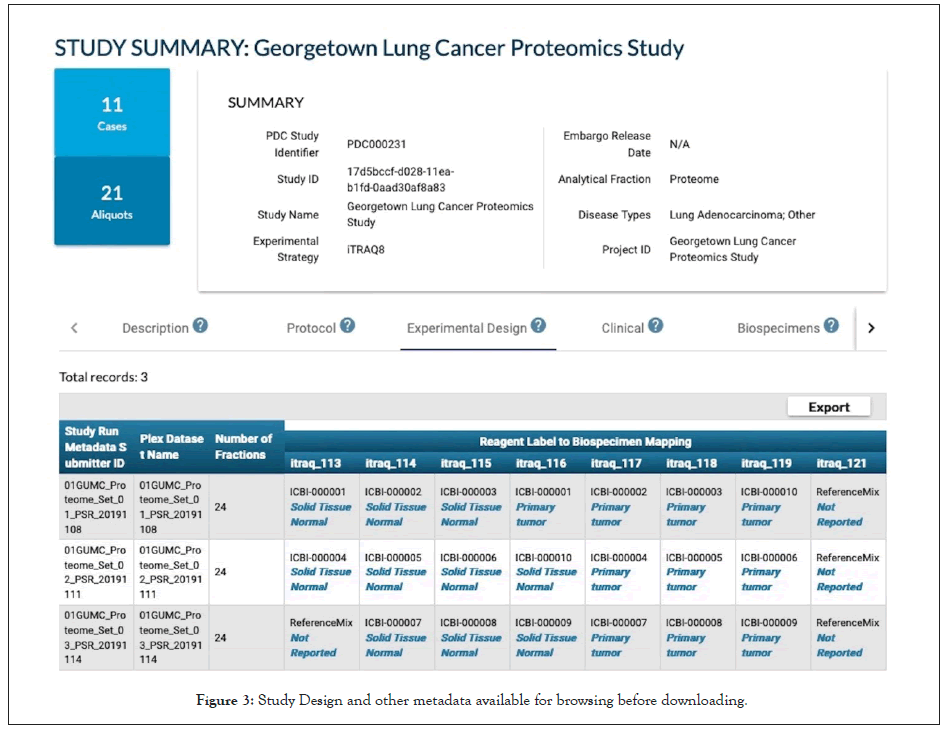
Figure 3: Study Design and other metadata available for browsing before downloading.
In summary the PDC data portal provides a comprehensive view of studies including the various metadata collected during the data submission, including biospecimen and clinical attributes, experimental design, even before downloading any files, Mapping and depositing data in the manner described here harmonizes your data to a common data model and community standards, making it possible to view the data alongside other high value studies cancer studies available in the PDC such as those from CPTAC and the International Cancer Proteogenomic Consortium (ICPC). This facilitates cross-study and cross-cancer queries to investigate questions about protein expression across cancer studies in PDC. The PDC enhances integration with other multi-omic data resources such as imaging TCIA and genomic GDC data; and, allows analysis using NCI analytic resources such as Seven Bridges without the need for data or tool transfer as these cloud resources can directly access PDC files in Amazon S3 buckets. Sharing standardized data is important and while it takes some effort it greatly enhances multi-disciplinary and collaborative research efforts.
Funding for GUMC/ LCCC: The Proteomics Shared Resource, the Histopathology and Tissue Shared Resource and the Innovation Center for Biomedical Informatics are partly supported by Lombardi Comprehensive Cancer Center Support Grant NCI P30-CA051008.
Funding for ESAC: Funded by NCI contract GS-35F-0539X_ HHSN261201700175U to ESAC, Inc.
The authors of this work have no competing interests to declare.
SM Conceptualization, Funding acquisition. PM Supervision, Writing - Original Draft, Data Curation, Investigation. RT Project administration. Data Curation. JM and CW Methodology, Investigation. SK Data Curation. KC and DB Resources. AF, DS, AB and PR Investigation, Validation. All authors Writing - Review & Editing.
The data underlying this article are available in the NCI Proteomics Data Commons with the unique study number PDC000231, https://pdc.cancer.gov/pdc/study/PDC000231.
Citation: McGarvey P, Thangudu RR, Ma J, Wu CI, Kannattikuni S, Chaldekas KM, et al. (2021) Sharing Data from an Academic Cancer Center Biospecimen and Proteomic Core Facilities Through the Proteomics Data Commons. J Proteomics Bioinform. 14:529.
Received: 05-Mar-2021 Accepted: 19-Mar-2021 Published: 26-Mar-2021 , DOI: 10.35248/0974-276X.21.14.529
Copyright: © 2021 McGarvey P, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Sources of funding : Funding for GUMC/ LCCC: The Proteomics Shared Resource, the Histopathology and Tissue Shared Resource and the Innovation Center for Biomedical Informatics are partly supported by Lombardi Comprehensive Cancer Center Support Grant NCI P30-CA051008. Funding for ESAC: Funded by NCI contract GS-35F-0539X_HHSN261201700175U to ESAC, Inc.