Drug Designing: Open Access
Open Access
ISSN: 2169-0138
ISSN: 2169-0138
Research Article - (2023)Volume 12, Issue 4
Background: Aspartic proteases possess catalytic sites for the hydrolysis of peptide bonds, which makes them potential drug targets in malaria parasites. Inhibiting Histo-Aspartic Protease (HAP), Aspartate (Asp215) and Histidine (His32) residues of P. falciparum disrupt the growth phase and ability to catalyze erythrocyte hemoglobin degradation.
Objectives: To synthesize compound 2-(2-benzoyl-4-methyl phenoxy) quinoline-3-carbaldehyde, through sp2 C-H activation protocols. To carry out in silico screening of fifty hypothetical compounds for their toxicity, pharmacokinetics, bioactivity score and binding affinities using Protox II web server, to carry out virtual screening of their toxicity and compliance with all drug-likeness rules. To carry out a molecular docking study of the docking of the ligands and ten references antimalarial drugs against HAP.
Methods: 2-(2-Benzoyl-4-methylphenoxy) quinoline-3-carbaldehyde was synthesized. In silico screening using Protox II webserver and molecular docking of the ligands and ten reference antimalarial drugs against HAP were carried out using ADME predictions and PyRx 0.8 AutoDock Vina Wizard.
Results: Nine lead compounds showed no toxicity to human cells. The lead compounds were generally highly or moderately bioactive for six bioactivity score parameters. Compound A31 was the best reference drug. While compound A31 and mefloquine both showed no interactions with either Asp215 or His32 in the binding pockets, compound A5 showed π-π stacking interactions. There was a significant hydrophobic interaction to suggest good water-lipid cell membrane transport within the Pf HAP protein, while the quinoline core exposure to a large solvent accessibility surface predisposes it to a more open conformation and binding interaction with the reactive site target residues.
Conclusion: Based on the other drug-likeness parameters investigated, compound A5, 2-(2-benzoyl-4-methylphenoxy)-7-methylquinoline-3-carbaldehyde, can be recommended as a possible candidate for new antimalarial drug development in line with SDG goal 3 on health and well-being.
Malaria; Mefloquine; Binding energy; Drug leads; Oral bioactivity score; Pharmacokinetics; Drug-likeness parameters
Malaria remains an important infectious disease characterized by acute febrile illness. According to the WHO, the global estimated burden is between 200 million and 300 million, with approximately 627,000 deaths in 2020. Among all the Plasmodium species causing malaria, Plasmodium falciparum is the most dreadful and prevalent in Africa. Over the past two decades, drug-resistant strains of P. falciparum have emerged to a distressing state, while also observing a reduction in the efficacy of currently available drugs. These pervasive challenges have motivated the search for novel drugs or redesigning existing chemical compounds as part of a more viable treatment protocol to engender sustainable global public health [1].
An emerging approach to overcome drug resistance shown by a disease pathogen such as the malaria parasite is to explore another biological component different from the traditional target sites. Plasmepsins, the aspartic proteinase that is present in malaria parasites, have become promising drug targets for malaria treatment. Histo-Aspartic Protease (HAP) is one of the four plasmepsins that resides in the food vacuole of P. falciparum. During the growth phase of Plasmodium, the parasite uses HAP to catalyze the degradation of erythrocyte hemoglobin at peptide bonds (known to be cleavage sites in the degradation pathway) to obtain amino acids for protein nutrient enrichment, whereas other plasmepsins are involved in other functions (Figure 1) [2].
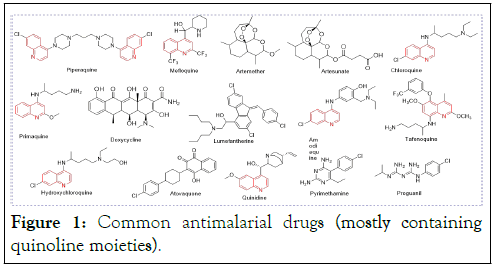
Figure 1: Common antimalarial drugs (mostly containing quinoline moieties).
Intuitively, taking cognizance of the Aspartate (Asp215) and Histidine (His32) residues within the active site of HAP and the high affinity for aspartic protease inhibitor Pepstatin-A may present a viable strategy to block site functionality, thereby inhibiting the protease of the Plasmodium parasite. Thus, knocking out HAP’s hemoglobin degrading ability could reduce propagation of the parasite in the host cells while preserving the hemoglobin of the infected erythrocytes [3].
Quinoline is an important organic compound that occurs in some natural compounds, especially alkaloids and pharmacologically active substances. Derivatives of the quinoline moiety have been reported to inhibit proteases of Plasmodium in vitro and in vivo. The quinoline moiety occurs in some current standard drugs shown in Figure 1, similar to compounds (Figure 2), for example, those used in the treatment of conditions such as antibacterial, anthelmintic, anticancer, antifungal, antihypertensive, anti-inflammatory, analgesic, and antiviral properties [4].
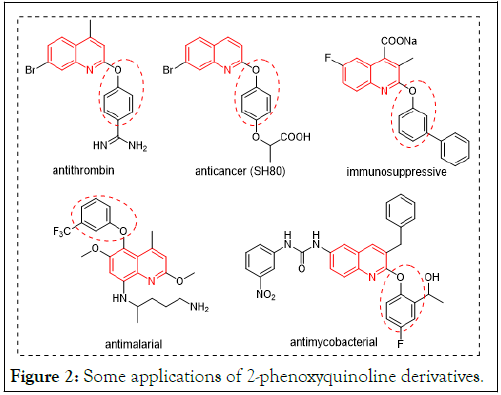
Figure 2: Some applications of 2-phenoxyquinoline derivatives.
There is a dearth of information on the drug-targeted design of quinoline derivatives as HAP inhibitors. Hence, we evaluated the in silico relative binding affinity of some hypothetical quinoline derivatives with the Histone-Aspartic Protease (HAP) of P. falciparum. We chose HAP because of its availability as a divergent vacuolar plasmepsin, with no similarity in any known species of Plasmodium [5]. Thus, we prepared 2-(2-benzoyl-4-methylphenoxy) quinoline-3-carbaldehyde (Figure 3) and evaluated it’s in silico toxicity, which returned as mildly carcinogenic (Table 1). Thereafter, fifty hypothetical derivative compounds were designed (Figure 4) for in silico screening to identify lead antimalarial candidates based on Absorption, Distribution, Metabolism, Excretion and Toxicity (ADMET) tests [6].
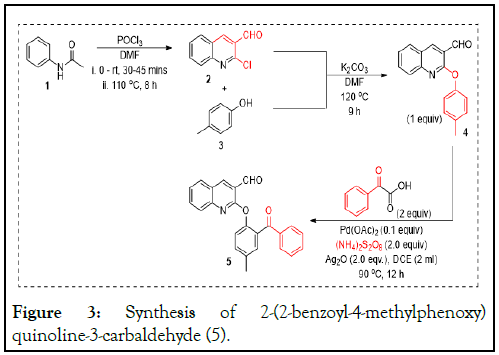
Figure 3: Synthesis of 2-(2-benzoyl-4-methylphenoxy) quinoline-3-carbaldehyde (5).
We further investigated their pharmacokinetics, bioactivity scores and molecular docking into the binding pockets of the histo-aspartic protease of Plasmodium falciparum. This drug-design protocol provides insights into the HAP enzyme active site amino acid residue binding interactions with the ligand compound, which is quite significant in the development of desirable drug-likable candidate inhibitors [7].
Synthesis of 2-(2-benzoyl-4-methylphenoxy quinoline-3-carbaldehyde
Phosphorous Oxychloride, POCl3 (28 ml), was added dropwise at 0°C in an ice bath to Dimethylformamide, DMF, and an orange color change was observed. This mixture was then added to acetanilide (10 g, 1 equiv.) dissolved in DMF (30 ml). The reaction temperature was increased from 0°C to 80°C for 9 hours while monitoring with TLC. At completion, the reaction mixture was allowed to cool and thereafter was poured dropwise into a stirring ice bath over 30 minutes. The precipitate obtained was washed with distilled water (50 ml × 5) to remove any remaining acid. The precipitate was filtered and air-dried over 48 hours [8].
To a mixture of p-cresol (1.70 ml, 10.44 mmol) and K2CO3 (4.33 g; 31.32 mmol) in DMF (15 ml) was added the precipitate; 2-chloroquinoline-3-carbaldehyde 2 (2.00 g, 10.44 mmol). The reaction mixture was stirred at 90°C for 9 hours. At completion, water (15 ml × 3) was added to the reaction mixture, and the solid obtained was filtered before recrystallization from ethyl acetate (10 ml) to afford a white solid, 2-phenoxyquinoline-3-carbaldehyde 4, which was used as a precursor for the benzoylation reaction.
In step 3, to a mixture of 2-phenoxychloroquinoline-3-carbaldehyde 4 (0.10 g; 0.38 mmol; 1.0 equiv.) placed in an oven-dried reaction tube was added (NH4)2S2O8 (0.24 g; 2.0 equiv.), Ag2O (1.0 eqv.) as a co-oxidant and Pd (OAc)2 (0.008 g; 0.04 mmol; 0.1 eqv.) as the catalyst. The mixture was flushed with nitrogen to evacuate air. Dichloroethane (2 ml) was added to the reaction and stoppered. The mixture was stirred at 100°C for 12 hours and monitored with TLC. After completion, ethyl acetate (20 ml × 3) was used for extraction. The organic layer was washed with water (15 ml × 3), dried with anhydrous sodium sulfate, filtered and concentrated under vacuum. The residue obtained was purified through column chromatography on silica gel (60-120 mesh) using hexane: Ethyl acetate (1:19) to give compound 5 (2-phenoxyquinoline-3-carbaldehyde) in 60% yield. The product was characterized by FTIR, 1H NMR and HRMS [9].
Toxicity prediction of compound
Toxicity tests of synthesized compound 5 were carried out by uploading its SMILES into the Protox II webserver. The hepatoxicity, carcinogenicity, immunotoxicity, mutagenicity and cytotoxicity data generated were extracted [10].
Preparation of its hypothetical compounds (A1-A50 and antimalarial reference drugs
Fifty (50) hypothetical compounds were conceptualized and drawn using Chemdraw 14.0, saved in.sdf format and their SMILES uploaded into the Protox II webserver (Figure 6). Ten antimalarial drugs, artesunate, doxycycline, tafenoquine, amodiaquine, artemeter, lumefantrine, primaquine, piperaquine, mefloquine and chloroquine, were obtained from PubChem for comparison. The downloaded structures were also saved in.sdf format, and their smiles were uploaded into the Protox II webserver for virtual screening to investigate their toxicity and compliance with all drug-likeness rules [11].
Selection of the Histone-Aspartic Protease (HAP) protein receptor
The crystal structure of the Histone-Aspartic Protease (HAP) protein molecule with a resolution of 2.10 Å was obtained from the protein data bank at rcsb.org in pdb format and prepared using BIOVIA discovery studio DS 2020 to remove unwanted ligands and water molecules. Polar hydrogen atoms were added accordingly [12].
In silico drug-likeness and ADME predictions
Drug-likeness analysis of compounds A1-A50 was undertaken using admetSAR2 to predict Adsorption, Distribution, Metabolism and Excretion (ADME) parameters for drug leads. The smiles were loaded into the webserver, and the results generated were extracted and analysed.
Bioactivity score
Bioactivity scores, such as G-Protein-Coupled Receptor (GPCR) ligand, Ion Channel Modulator (ICM), Kinase Inhibitor (KI), Nuclear Receptor Ligand (NRL), Protease Inhibitor (PI) and enzyme inhibitor EI, of the lead compounds identified were evaluated using the online Molinspiration drug-likeness score at www.molinspiration.com. This was done by loading the smiles of the lead compounds into the web server. The values obtained were plotted using excel [13].
Molecular docking study
Investigation of the inhibitory ability of synthesized compound 5 and selected hypothetical compounds was carried out against Histo-Aspartic Protease (HAP) using docking simulations performed with PyRx 0.8 AutoDock Vina Wizard before conversion to AutoDock macromolecules. Flexible ligand to rigid protein procedures was followed and all potential binding sites on the target protein were utilized. Docking was performed within a 90 × 75 × 60 cubic grid centered on the protein and enclosing the entire protein, which lasted for approximately one hour. A grid spacing of 1.00 Å was used for the calculation of the grid maps using the autogrid module of Autodock tools using a set of nine (9) independent runs for each ligand [14].
The potential binding sites identified influence the selected energetically favorable binding conformations using AutodockVina. The mode, binding affinity of the modes and RSB (upper and lower) values were obtained as guides to the highest binding score for each ligand. The mode with the best binding affinity was selected. Analysis of the ligand-protein complex was performed using DS visualizer. All software was run on PC-based machines running Microsoft windows 10 operating systems.
Synthesis of 2-(2-benzoyl-4-methylphenoxy) quinoline-3-carbaldehyde (5) and preparation of hypothetical compounds (A1-A50) as ligands
Compound 5, 2-(2-benzoyl-4-methylphenoxy) quinoline-3-carbaldehyde, was obtained as shown in scheme 1 and characterized using FTIR, HRMS and 1H-NMR. We investigated the drug-likeness of compound 5, which showed mild carcinogenicity (Table 1). To identify candidates with better drug-likeness, fifty hypothetical compounds were designed based on structural modifications with substituents at positions 4, 5, 6, 7, and 8 of the quinoline core, the Meta position of the methylphenoxy group and the para position of the benzoyl group (Figure 4) [15].
Structural elucidation of compound: White solid; reaction time: 12 h; yield: 60%; m.pt: 119°C-121°C; IR (neat) v max (cm-1) 3057, 2922, 2856, 2739, 1754, 1690, 1612, 1590, 1494, 1461, 1343, 1257, 1199, 1097, 760; 1H NMR (400 MHz, CDCl3) δ 9.75 (s, 1H), 8.32 (s, 1H), 7.97 (d, J=8.3 Hz, 1H), 7.82 (d, J=8.2 Hz, 1H), 7.80 (t, J=8.2 Hz, 1H), 7.74 (t, J=7.5 Hz, 1H), 7.61 (d, J=8.2 Hz, 1H), 7.51 (d, 1H), 7.39 (t, 1H), 7.19 (s, 1H), 6.86 (d, 1H), 6.84 (d, 1H), 2.41 (s, 3H). HRMS (ESI): Calc. for [(C24H17NO3)] (M+H)+368.1281, found 368.1283.
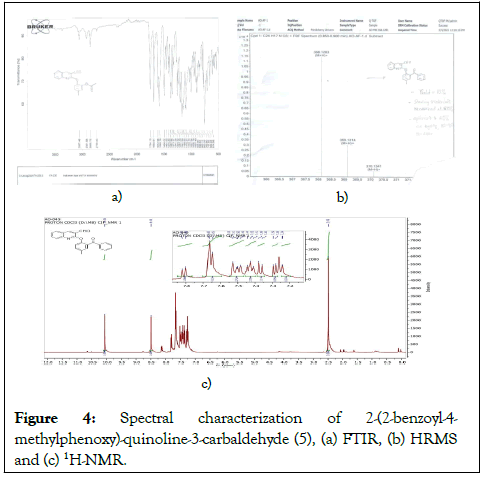
Figure 4: Spectral characterization of 2-(2-benzoyl-4-methylphenoxy)-quinoline-3-carbaldehyde (5), (a) FTIR, (b) HRMS and (c) 1H-NMR.
Toxicity results of compound (5), hypothetical compounds (A1-A50) and ten antimalarial reference drugs
All fifty hypothetical compounds (A1-A50) in Figure 4 and ten reference drugs, artesunate, doxycycline, tafenoquine, amodiaquine, artemeter, lumefantrine, primaquine piperaquine, mefloquine and chloroquine, were virtually investigated for their toxicity profiles, including hepatoxicity, carcinogenicity, immunogenicity, mutagenicity and cytotoxicity, as given in Table 1 (Figure 6). Noticeably, while compound 5 and forty-one of the hypothetical derivatives failed one or more of those tests suggesting possible toxic and carcinogenic activities, nine (9) lead compounds A5, A20, A31, A33, A34, A36, A45, A48 and A49 (Figure 5) were fully compliant (Table 1) [16].
Figure 5: Hypothetical fifty compounds A1-A50 obtained from structural modifications of compound (5).
| Target | Compound 5 | A5 | A31 | A36 | A20 |
|---|---|---|---|---|---|
| Hepatotoxicity | Inactive (0.56) | Inactive (0.97) | Inactive (0.50) | Inactive (0.52) | Inactive (0.54) |
| Carcinogenicity | Active (0.54) | Inactive (0.57) | Inactive (0.52) | Inactive (0.52) | Inactive (0.54) |
| Immunotoxicity | Inactive (0.83) | Inactive (0.99) | Inactive (0.82) | Inactive (0.71) | Inactive (0.67) |
| Mutagenicity | Inactive (0.55) | Inactive (0.85) | Inactive (0.56) | Inactive (0.56) | Inactive (0.60) |
| Cytotoxicity | Inactive (0.75) | Inactive (0.78) | Inactive (0.74) | Inactive (0.75) | Inactive (0.75) |
|
|
|
|
|
|
|
| Hepatotoxicity | Inactive (0.60) | Inactive (0.60) | Inactive (0.52) | Inactive (0.54) | Inactive (0.54) |
| Carcinogenicity | Inactive (0.56) | Inactive (0.56) | Inactive (0.51) | Inactive (0.41) | Inactive (0.54) |
| Immunotoxicity | Inactive (0.65) | Inactive (0.62) | Inactive (0.67) | Inactive (0.69) | Inactive (0.60) |
| Mutagenicity | Inactive (0.53) | Inactive (0.53) | Inactive (0.52) | Inactive (0.60) | Inactive (0.60) |
| Cytotoxicity | Inactive (0.71) | Inactive (0.71) | Inactive (0.76) | Inactive (0.75) | Inactive (0.75) |
Table 1: Toxicity prediction and probability values of lead compounds.
Figure 6: Nine nontoxic hypothetical derivatives of compound 5.
Interestingly, the toxicity results for the reference drugs, as presented in Table 2, returned only mefloquine as compliant, while others had one or more violations in comparison with the nine lead compounds. Mefloquine was therefore selected for further virtual study along with the selected lead compounds [17].
| Target | Mefloquine | Piperaquine | Artesunate | Doxycycline | Tafenoquine |
|---|---|---|---|---|---|
| Hepatotoxicity | Inactive (0.75) | Inactive (0.78) | Inactive (0.76) | Active (0.54) | Inactive (0.78) |
| Carcinogenicity | Active (0.76) | Inactive (0.71) | Inactive (0.65) | Inactive (0.77) | Inactive (0.63) |
| Immunotoxicity | Inactive (0.84) | Active (0.93) | Active (0.87) | Active (0.99) | Active (0.99) |
| Mutagenicity | Inactive (0.68) | Active (0.50) | Inactive (0.63) | Inactive (0.95) | Active (0.54) |
| Cytotoxicity | Inactive (0.74) | Inactive (0.82) | Inactive (0.87) | Inactive (0.90) | Inactive (0.63) |
|
|
|
|
|
|
|
| Hepatotoxicity | Inactive (0.61) | Inactive (0.77) | Inactive (0.70) | Inactive (0.84) | Inactive (0.90) |
| Carcinogenicity | Active (0.61) | Inactive (0.66) | Inactive (0.61) | Inactive (0.59) | Inactive (0.66) |
| Immunotoxicity | Active (0.99) | Active (0.92) | Active (0.99) | Active (0.99) | Active (0.69) |
| Mutagenicity | Inactive (0.75) | Inactive (0.60) | Inactive (0.60) | Active (0.79) | Active (0.94) |
| Cytotoxicity | Inactive (0.53) | Inactive (0.94) | Inactive (0.67) | Inactive (0.61) | Inactive (0.93) |
Table 2: Toxicity prediction and probability values of standard drugs against P. falciparum.
In silico drug-likeness and ADME predictions
Our results in Table 3 show that the lead compounds have hydrogen bond donors (nitrogen-hydrogen and oxygen-hydrogen bonds (0-1), and (<5), while the hydrogen bond acceptors (all nitrogen or oxygen atoms) (4-7) also (<10), are in compliance with the Rule of Five (RO5). Their molecular weights range from 367.40 g/mol to 449.40 g/mol, which also fit the 150 g/mol to 500 g/mol rule. The TPSA observed was 56.26 Å2 to 104 Å2, which falls within the range of 20 Å2 to 130 Å2. The number of rotatable bonds was not more than 9 [18].
Compounds A20, A45 and A49 were less permanent than the others, as revealed by their higher negative log Kp values (-5.13 to -5.18). Unlike others, compounds A31, A36 and A45 showed low gastrointestinal absorption values. This could be attributed to the presence of the trifluoromethyl group at position 6 of compound A31, the thiol group at position 8 of compound A36, and both methoxy at position 6 and thiol at position 3 of the methylphenoxyl ring. Interestingly, compound 5 is blood brain barrier permeant, unlike all the lead compounds. Compounds A31, A33, A34 and A36 are P-gp substrates.
| Physicochem* | 5 | A5 | A31 | A36 | A20 | A33 | A34 | A45 | A48 | A49 |
|---|---|---|---|---|---|---|---|---|---|---|
| MW | 367.4 | 381.42 | 449.42 | 427.51 | 383.4 | 409.48 | 409.48 | 429.49 | 383.4 | 383.4 |
| #Rot_b | 5 | 5 | 6 | 7 | 5 | 6 | 6 | 6 | 5 | 5 |
| #HA | 4 | 4 | 7 | 4 | 5 | 4 | 4 | 5 | 5 | 5 |
| #HD | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 |
| TPSA | 56.26 | 56.26 | 56.26 | 81.56 | 76.49 | 56.26 | 56.26 | 104.29 | 76.49 | 76.49 |
| N_atoms | 28 | 29 | 33 | 31 | 29 | 31 | 31 | 31 | 29 | 29 |
| N_viol | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Volume | 330.21 | 346.77 | 378.07 | 381.7 | 383.23 | 380.16 | 380.16 | 373.42 | 383.23 | 338.23 |
| Log Kp | -4.83 | -4.66 | -4.45 | -4.58 | -5.18 | -4.29 | -4.29 | -5.13 | -4.79 | -5.18 |
| Bioav | 0.55 | 0.55 | 0.55 | 0.55 | 0.55 | 0.55 | 0.55 | 0.55 | 0.55 | 0.55 |
| GI ab | High | High | Low | Low | High | High | High | Low | High | High |
| BBB perm | Yes | No | No | No | No | No | No | No | No | No |
| Pgp sub | No | No | Yes | Yes | No | Yes | Yes | No | No | No |
| Note: *TPSA=Total Polar Surface Area; natoms=number of atoms in the molecule; MW=Molecular Weight; nON=number of hydrogen bond acceptors; nOHNH=number of hydrogen bond donors; nviol=number of violations; nrotb=number of rotatable bonds. | ||||||||||
Table 3: Physicochemical properties and drug-likeness of compound 5 and the leads.
Six of the nine derivatives in addition to compound 5 exhibited high gastrointestinal absorption, except A31, A36 and A45. Furthermore, unlike all the other lead compounds, compound 5 can penetrate the Blood Brain Barrier (BBB) (Table 4). The nine lead compounds all have good oral bioavailability of 0.55, although with one allowable violation. Inhibition of cytochrome P450 isoenzymes has been linked to the major cause of pharmacokinetics related drug-drug interactions, which could lead to toxic or adverse effects when there is lower clearance or accumulation of drugs or metabolites. Table 4 reveals that the lead compounds are CYP2C19 inhibitors that are substrates of CYP2D6. These cytochromes are involved in the metabolism and elimination of approximately 25% of clinically used drugs involved in the addition or removal of certain functionalities through hydroxylation, demethylation and dealkylation [19].
| Derivative | Lipinski | Ghose | Veber | Egan | Muegge | CYP2C19 | CYP2D6 |
|---|---|---|---|---|---|---|---|
| 5 | 0 | 0 | 0 | 0 | 1 | Yes | No |
| A5 | 0 | 1 | 0 | 0 | 1 | Yes | No |
| A31 | 1 | 1 | 0 | 1 | 1 | Yes | No |
| A36 | 0 | 1 | 0 | 1 | 1 | Yes | No |
| A20 | 0 | 0 | 0 | 0 | 0 | Yes | No |
| A33 | 0 | 1 | 0 | 1 | 1 | Yes | No |
| A34 | 0 | 1 | 0 | 1 | 1 | Yes | No |
| A45 | 0 | 1 | 0 | 0 | 1 | Yes | No |
| A48 | 0 | 0 | 0 | 0 | 1 | Yes | No |
| A49 | 0 | 0 | 0 | 0 | 0 | Yes | No |
Table 4: Drug violations and cytochrome inhibition ability of compound 5 and the leads.
Bioactivity score
The probability of drug leads as potential candidates can be evaluated using their bioactivity scores.
In Figure 7, all the lead compounds generally fall within high or moderate bioactivity for all the parameters. Specifically, compounds A20 and A48 are highly active for five of the six parameters with bioactivity scores from 0.00 to 0.33. Compounds A5 and A31 have high bioactivity as kinase inhibitors (0.23 and 0.22) with the capacity to block cancer cells, as nuclear receptor ligands (0.18 and 0.26) for hydrophobic molecules such as fatty acids, cholesterol and lipophilic hormones, and as glycoprotein receptors GPCR (0.07 and 0.08), which regulate metabolic enzymes or promoter proteins, while A5 is also an enzyme inhibitor with a value of 0.1 that is able to bind to other available sites on the enzyme. The compound with the lowest bioactivity score is A36. All the lead compounds are only moderately active as protease inhibitors with values of (-0.18 to -0.28), implying that they have the capacity to prevent new HIV cells from becoming mature virus. Only compounds A20 and A48 are highly active as ion channel modulators with values of 0.04 and 0.07, respectively, which are above 0.00 (Table 5) [20].
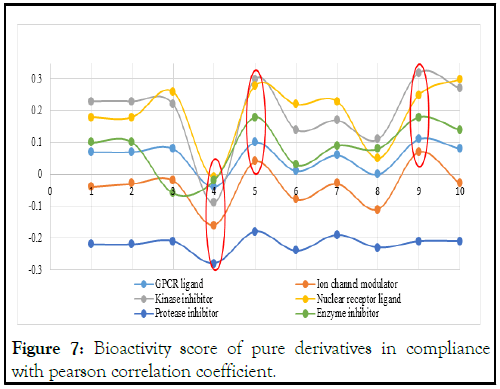
Figure 7: Bioactivity score of pure derivatives in compliance with pearson correlation coefficient.
| Bioactivity score of pure derivatives | 5 | A5 | A31 | A36 | A20 | A33 | A34 | A45 | A48 | A49 |
|---|---|---|---|---|---|---|---|---|---|---|
| Number code of derivatives | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
Table 5: Bioactivity score of pure derivatives in compliance with pearson correlation coefficient.
Molecular docking study
The results from the docking of ligands and reference drugs against HAP are presented in Table 6. The binding energies for compound A31 (-11.3 kcal/mole) and compound A5 (-11.2 kcal/mole) are higher than that for compound 5 (-10.9 kcal/mol). Additionally, the next six of the lead compounds have binding energies of (-10.8 kcal/mole to -9.8 kcal/mole) all higher than the ten reference drugs investigated, for which the best performing mefloquine had -9.6 kcal/mole while the lowest was chloroquine with -6.0 kcal/mole (Figure 8) [21].
| 9Ligand | Binding affinity |
|---|---|
| A31_uff_E=300.34 | -11.3 |
| A5_uff_E=278.30 | -11.2 |
| A1/5_uff_E=277.56 | -10.9 |
| A20_uff_E=282.50 | -10.8 |
| A33_uff_E=300.83 | -10.5 |
| A49_uff_E=284.88 | -10.5 |
| A48_uff_E=343.12 | -9.9 |
| A36_uff_E=283.54 | -9.8 |
| A45_uff_E=414.27 | -9.8 |
| Mefloquine | -9.6 |
| Piperaquine | -9 |
| A34_uff_E=340.75 | -8.7 |
| Artesunate | -8.5 |
| Doxycycline | -8.5 |
| Tafenoquine | -8.5 |
| Amodiaquine | -8.4 |
| Artemeter | -8.3 |
| Lumefantrine | -7.3 |
| Primaquine | -6.9 |
| Chloroquine | -6 |
Table 6: Binding energies of pure derivatives and reference drugs.
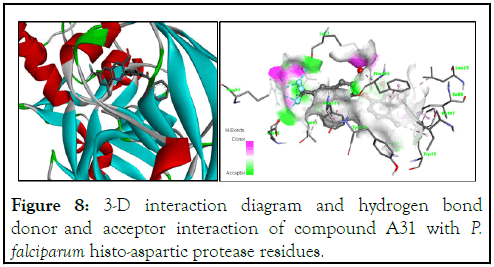
Figure 8: 3-D interaction diagram and hydrogen bond donor and acceptor interaction of compound A31 with P. falciparum histo-aspartic protease residues.
The 3-D structure of P. falciparum histo-aspartic protease (PDB ID: 3QVC) and its hydrogen acceptor/donor surface interaction with compound A31 is shown in Figure 9. We observed in the 2-D view (Figure 9) the hydrogen binding interaction of the carbaldehyde oxygen atom of the quinoline core with Phe109 at a bond length of 3.51 Å. Additionally, strong fluorine bonds of the trifluoromethyl groups at position 7 with Glu86, Arg91, Lys7 and Ala10 (bond lengths ranging from 2.98 Å to 3.58 Å) may presumably be responsible for the high inhibitory interactions of 2-(2-benzoyl-4-methylphenoxy)-8-methyl-6-(trifluoromethyl)quinoline-3-carbaldehyde (A31) with Plasmodium falciparum (HAP). Other bonds contributing to the interaction, as presented in Figure 9, are alkyl, π-π alkyl, π-π stacked and π alkyl at various bond lengths with the residues of the protein [22].
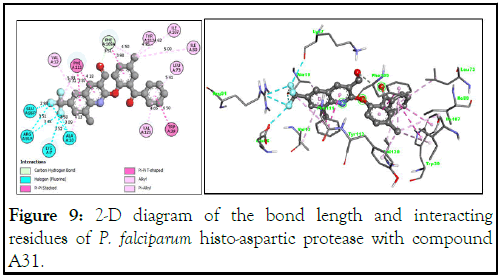
Figure 9: 2-D diagram of the bond length and interacting residues of P. falciparum histo-aspartic protease with compound A31.
Hydrophobic interactions are highly crucial for the folding of proteins, especially in keeping the protein stable and biologically active through a decrease in surface area, thereby reducing undesirable interactions with water. Herein, compound A31 exhibits hydrophobic interactions with the P. falciparum HAP amino acid residues, especially the 2-benzoyl-4-methylphenoxy side of the molecule interacting with Leu73, Ile80, Tyr112, Phe111, Trp39 and Ile107.
Interaction with the trifluoromethyl side of the quinoline molecule with Glu86, Arg91, Lys7 and Ala10 clearly appears to be hydrophilic due to the electronegative character of the fluorine atom, as shown in Figure 10. Solvent accessibility (SAS) is a key feature of proteins for determining the folding and stability of a molecule. In Figure 10, the solvent accessibility surface and blue region in the 3D interaction are large, thereby suggesting a better interaction of compound A31 with the binding pocket of the HAP protein.
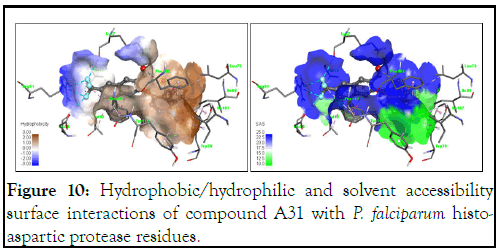
Figure 10: Hydrophobic/hydrophilic and solvent accessibility surface interactions of compound A31 with P. falciparum histo-aspartic protease residues.
Similarly, the 3-D structure for compound A5 is shown in Figure 11 with a binding energy of -11.2 kcal/mol. The absence of hydrogen bonds does not reduce its efficacy as a HAP inhibitor due to other interactions, such as π-cation (pi electrons of the quinoline core and the amino hydrogen of the side chain of Lys7), alkyl and π-alkyl (ligand and amino acid residues such as Val120, Leu73, Tyr410, Leu73, Ile80, Ile107 and Pro110), π-π stacking and π-π T-shaping (compound A5 and Phe111, Phe109, His32 and Trp39), all contributing to its high binding energy (Figure 12).
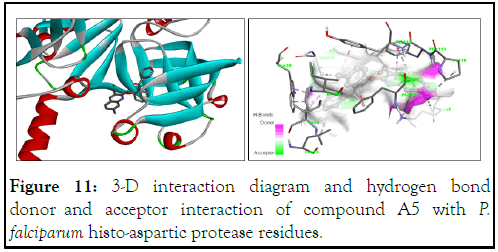
Figure 11: 3-D interaction diagram and hydrogen bond donor and acceptor interaction of compound A5 with P. falciparum histo-aspartic protease residues.
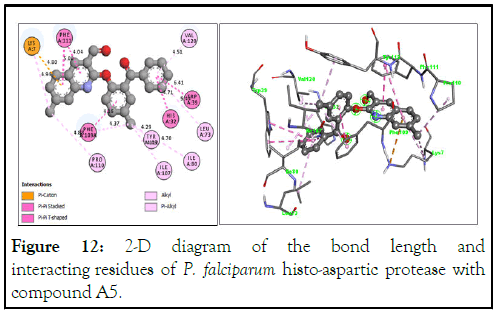
Figure 12: 2-D diagram of the bond length and interacting residues of P. falciparum histo-aspartic protease with compound A5.
We observed in Figure 13 that compound A5 displays significant hydrophobic interactions with all observed binding residues except Lys7, suggesting good water-lipid interface transport between the cell membrane of the Pf HAP protein. Furthermore, the quinoline core of compound A5 is exposed to better solvent accessability, implying a more open conformation predisposed to easier interaction with the reactive sites of the target residues (Figure 14).
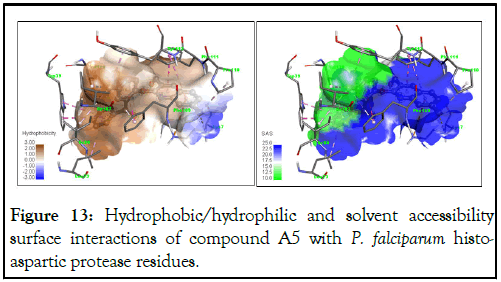
Figure 13: Hydrophobic/hydrophilic and solvent accessibility surface interactions of compound A5 with P. falciparum histoaspartic protease residues.
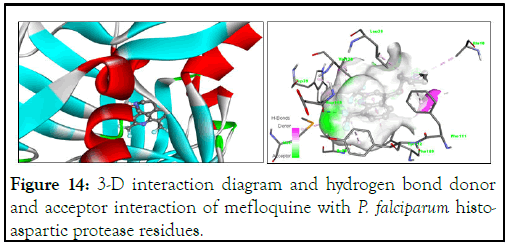
Figure 14: 3-D interaction diagram and hydrogen bond donor and acceptor interaction of mefloquine with P. falciparum histo-aspartic protease residues.
The binding interactions of the best reference drug, mefloquine, with P. falciparum histo-aspartic protease residues (Figure 15) showed that the trifluoro groups attached to position 6 in compound A31 and position 8 in mefloquine both appear to contribute to their increased activity. However, unlike compound A31, mefloquine does not have any hydrogen bonding, which could have explained its lower binding energy.
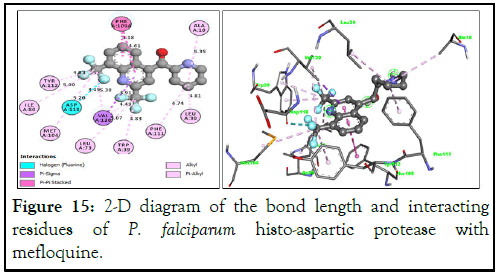
Figure 15: 2-D diagram of the bond length and interacting residues of P. falciparum histo-aspartic protease with mefloquine.
Similarly, the solvent accessibility surface interaction of compound A31 is higher for mefloquine, suggesting a better interaction in the binding pocket of the HAP protein (Figure 16).
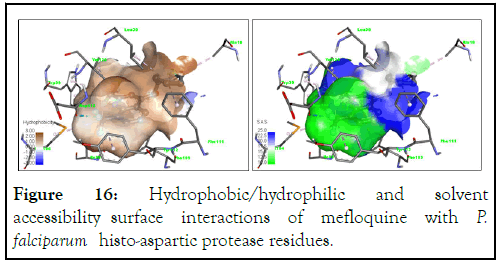
Figure 16: Hydrophobic/hydrophilic and solvent accessibility surface interactions of mefloquine with P. falciparum histo-aspartic protease residues.
In this study, through toxicity profile tests, we evaluated synthesized compound 5 and its fifty hypothetical compounds A1-A50 for their drug-likeness using virtual tools to identify lead drug candidates to address resistance to current reference antimalarial drugs. Nine hypothetical compounds showed no toxicity to human cells.
Compounds A5 and A31 have high bioactivity as kinase inhibitors, nuclear receptor ligands, and glycoprotein receptors GPCR. Additionally, compound A5 is an enzyme inhibitor able to bind to other available sites on the HAP enzyme.
We join others to hypothesize that since aspartate (Asp215) and histidine (His32) residues in the active site of HAP are desirable for parasite growth and particularly contribute to its virulence, ligand interaction at this location will serve as a good inhibitor in a drug-development protocol.
Herein, while compound A31 with a binding energy of -11.3 kcal/mole does not show evidence of interaction with either Asp215 or His32, compound A5 with a binding energy of -11.2 kcal/mole has π-π stacking interactions with His32. Mefloquine, the best performing reference drug, also does not show any interaction with either Asp215 or His32.
Furthermore, compound A5 was observed to show significant hydrophobic interactions with all observed binding residues except Lys7, suggesting good water-lipid interface transport between the cell membrane of the Pf HAP protein and exposure of the quinoline core to better solvent accessibility, which suggests that the conformation is more open, enabling better binding with the reactive sites of the target residues. Our study shows that compound A5 may be considered in a new drug development protocol for antimalarial disease.
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
The authors declare the no financial interests/personal relationships which may be considered as potential competing interests.
This work was funded, in parts, by the university of Lagos Central Research committee (CRC no. 2015/25), Nigerian government TetFund IBR.
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Oluwafemi Aina and Bello Adebayo. The first draft of the manuscript was written by Luqman Adams and Oluwole B. Familoni and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Google Scholar] [PubMed]
Citation: Oluwafemi AS, Luqman AA, Adebayo BJ, Oluwole FB (2023) Plasmodium falciparum Histo-Aspartic Protease (HAP) Inhibitor: Toxicity Investigation and Docking Study of 2-(2-benzoyl-4-methylphenoxy)quinoline-3-carbaldehyde Derivatives. Drug Des. 12:260.
Received: 21-Apr-2022, Manuscript No. DDO-23-23687; Editor assigned: 24-Apr-2023, Pre QC No. DDO-23-23687 (PQ); Reviewed: 08-May-2023, QC No. DDO-23-23687; Revised: 22-Jun-2023, Manuscript No. DDO-23-23687 (R); Published: 28-Dec-2023 , DOI: 10.35248/2169-0138.23.12.260
Copyright: © 2023 Oluwafemi AS, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.