Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research Article - (2021)Volume 14, Issue 2
Salinity is a major abiotic stress that adversely affects plant growth and development. Canola (Brassica napus L.) is an important oilseed crop in the world, and its yield decreases drastically with increasing salinity. To date, little is known about the molecular mechanisms underlying its salt stress response and tolerance. This study combines physiological assays with comparative proteomics to understand how B. napus plants respond to salt stress. The changes in relative water content, electrical conductance, stomata conductance, intercellular CO2 concentration, transpiration rate, photosynthesis rate, water usage efficiency, respiration rate, chlorophyll fluorescence, antioxidant enzyme activities, soluble sugar, proline and betaine in B. napus plants under different NaCl concentrations were analyzed. Proteomic profiles of B. napus plants under 100, 200 and 400 mM NaCl treatment at 7 day and 14 day were acquired using iTRAQ LC-MS/MS based quantitative proteomics. A total of 2316 proteins were identified in B. napus leaves, of which 614 proteins showed differential expression under salt stress. These proteins were mainly involved in 10 processes, of which proteins in stress and defense, metabolism and photosynthesis pathways ranked the top three. Subcellular localization analysis showed that most proteins were located in chloroplast, cytoplasm, mitochondria and nucleus. A total of 138 differentially expressed proteins were predicted to interact with each other. These results have provided a comprehensive view of the physiological and molecular processes taken place in B. napus leaves under salt stress, and revealed the molecular mechanisms underlying salt tolerance of B. napus plants.
Brassica napus; Salt stress; Physiological analysis; Proteomics; iTRAQ LC-MS/MS
APX: Ascorbate Peroxidase; AsA-GSH cycle: Ascorbateglutathione Cycle; ATPS: ATP Sulfurylases; CA: Carbonic Anhydrase; CAB: Chlorophyll a b-binding Protein; CAT: Catalase; Ci: Intercellular CO2 Concentration; Cond: Stomatal Conductance; CS: Cysteine Synthases; CYP: Chaperone/ Chaperonin Proteins; Cytb6f: Cytochrome b6-f Complex Protein; DEPs: Differentially Expressed Proteins; 2-DE: Two-dimensional Gel Electrophoresis; DW: Dry Weight; ER: Endoplasmic Reticulum; FNR: Ferredoxin NADP+ Reductase; FW: Fresh Weight; GADPH: Glyceraldehyde-3-phosphate Dehydrogenase; Gly I: Glyoxalase I; Gly II: Glyoxalase II; GO: Glycolate Oxidase; GPX: Guaiacol Peroxidase; GR: Glutathione Reductase; GSH: Reduced Glutathion; GS: Glutamine Synthetase; GST: Glutathione S-Transferase; HK: Histidine Kinases; HSPs: Heat Shock Proteins; iTRAQ: Isobaric Tagging for Relative and Absolute Quantification; MG: Methylglyoxal; NO: Nitric Oxide; OEE2: Oxygen-Evolving Enhancer Protein 2; OEC: Oxygen-Evolving Complex; PDI: Protein Disulfide-Isomerase; PGK: Phosphoglycerate Kinase; PKS5: PROTEIN KINASE5; PMA: Plasma Membrane ATPase; Pn: Photosynthesis Rate; POD: Peroxidase; POR: Protochlorophyllide Reductase; PPase: Pyrophosphatase; PPIase: Peptidyl-prolyl cis-trans Isomerase; PPIs: Protein-protein Interactions; PRK: Phosphoribulokinase Precursor; PrxR-Trx cycle: Peroxiredoxin-thioredoxin pathway; RCA: RubisCO Activase; REC: Relative Electrical Conductance; ROS: Reactive Oxygen Species; RPI: Ribulose-5-phosphate Isomerase; RR: Two-component System Response and Regulator; RubisCO: Ribulose 1,5-biphosphate Carboxylase/Oxygenase; RWC: Relative Water Content; S: Sulfur; Se: Selenite; SLG: S-lactoylglutathione; SOD: Superoxide Dismutase; SOS: Salt- Overly-Sensitive; TK: Transketolase; TPI: Triosephosphate Isomerase; Tr: Transpiration Rate; VHA: V-type H+-ATPase; WUE: Water-Use Efficiency
Plants are constantly challenged by various biotic and abiotic stresses during their life cycle, which negatively affect their growth, development and productivity [1,2]. Salinity as one of the major abiotic stresses affects more than 800 million hectares of land, equivalent to more than 6% of the earth area [3-5]. Approximately 400 million hectares of agricultural land are affected by salinity and this number keeps going up [6]. Therefore, salt stress poses a major global problem for agriculture. It has stimulated immense interest in elucidating salt tolerance mechanisms and developing strategies toward boosting crop salt tolerance [7,8].
Salt stress causes perturbation to many plants physiological and biochemical processes, including photosynthesis, nutrient and water uptake, root growth, and cellular metabolism [9-11]. Plant responses to salt stress include the following phases [1]: Salinity causes low water potential of roots, resulting in osmotic stress. Meanwhile, Na+ and Cl- ions are transported to the shoots, resulting in ionic stress. Subsequently, increased production of reactive oxygen species (ROS) leads to oxidative stress. The osmotic stress, ionic toxicity and oxidative stress cause deleterious effects to plants [12,13]. To survive salt stress, plants respond and adapt with sophisticated mechanisms that include developmental, morphological, physiological and biochemical strategies and require alterations in gene expression and changes in the protein profiles [7,14,15]. At physiological level, plants have developed cellular adjustment strategies to reduce salt stress damages, e.g., accumulating compatible solutes (soluble sugars, proline and betaine) [15,16], excluding Na+ and Cl− in roots [5] and scavenging of ROS [17]. The effectiveness of the plant antioxidant system can be measured by the activities of the ROS-scavenging enzymes such as superoxide dismutase (SOD), ascorbate peroxidase (APX), catalase (CAT), glutathione reductase (GR), guaiacol peroxidase (GPX) and peroxidase (POD) [18-21].
Many salt stress responsive genes in different plants have been identified [22,23]. For example, 1696 genes in rice (Oryza sativa) [24], 2696 genes from Soybean (Glycine max) [25], 7217 genes from Arabidopsis thaliana [26], 472 genes from maize (Zea mays) [27], 163 genes from Canola (B. napus) [28], 3310 genes from two genotypes of Medicago truncatula [29], and 840 genes from wheat (T. aestivum cv. Chinese Spring) [30] showed differential expression under salt stress. However, transcriptomic approaches may only partially contribute to the understanding of plant stress responses because many transcripts may undergo a number of posttranscriptional modifications, and even do not make proteins [7,31]. Proteomics, the large-scale study of proteins, has the potential to fill this gap. Proteins are “actor” molecules and a profound analysis of the proteome is essential to understanding the molecular mechanisms underlying plant salt stress response [1]. Recently, isobaric tagging for relative and absolute quantification (iTRAQ)- based quantitative proteomics approach [32,33] was used to identify salt responsive proteins in several plant species [14,34-40]. For example, 31 and 32 differentially expressed proteins (DEPs) in leaves of A. thaliana and Thellungiella halophila under salt stress were identified, respectively [14]. In rice shoots, 56 DEPs were identified after salt stress treatment [34], and in rice suspension cells 521 salt stress responsive proteins were identified [35]. In maize roots, 28 salt-responsive proteins were identified [36]. In tomato roots, 313 proteins responsive to NaCl and NaHCO3 were observed [37]. In sugar beet leaves and roots, 75 and 43 DEPs under salt stress were identified, respectively [38]. In the chloroplasts of Kandelia cande [39] and in the leaves of Tangut Nitraria [40], 76 and 71 DEPs were identified, respectively.
Canola (Brassica napus L.) belongs to the family of Brassicaceae. Canola is the third most important oilseed crop after palm and soybean, cultivated worldwide for oil production [41]. Canola oil, rich in polyunsaturated fatty acids, is considered a healthy ingredient. Canola is also considered as one of the essential sources for biodiesel fuel [42,43]. Like other important crops, environmental stresses reduce canola yield and production. Although physiological changes in Canola under salt stress have been studied [44-49], the studies did not include comprehensive physiological analyses to include relative water content, electrical conductance, photosynthesis, chlorophyll fluorescence, soluble sugar, proline, betaine contents and antioxidant enzyme activities at different time points under different levels of salt stress. To date, only a few proteomic studies in Canola under salt stress have been reported. Bandehagh et al. identified 46 proteins in canola leaves under salt stress (0, 175 mM and 350 mM NaCl) at one time point using 2-DE (two-dimensional gel electrophoresis) approach [50]. The single time point snapshot missed dynamic protein changes in the course of plant response to salt stress. Other 2-DE proteomics of canola salt stress responses only identified 42 [51] and 21 proteins [52]. It is well-known that the 2D gel approach is limited to mostly abundant proteins [53-55]. A recent gel-free proteomics on how Pseudomonas fluorescens bacteria improved canola tolerance to salt stress identified a couple of hundred proteins [56]. In addition, proteomic changes of canola under drought stress over a 14-day period were examined using iTRAQ LC-MS/MS. A total of 1976 proteins expressed during drought were identified, revealing changes of protein abundance and post-translational modifications (PTMs) [57].
In the present work, physiological and proteomic changes of B. napus in response to different levels of salt stress were profiled in a time course study. Different types of morphological and physiological data were obtained from B. napus seedlings treated with 0, 50 mM, 100 mM, 200 mM and 400 mM NaCl for different periods of time. Then an iTRAQ-based quantitative proteomics approach was used to identify and quantify DEPs in the leaves under 100 mM, 200 mM and 400 mM NaCl treatments for 7 days and 14 days, respectively. This is a largescale study using iTRAQ 2D LC-MS/MS in profiling proteomic changes in leaves of canola plants under salt stress. The results have enhanced understanding of B. napus salt-responsive proteins and their potential functions, and provided a comprehensive view of the physiological and molecular processes in the leaves under salt stress, which lead to deeper insights into the molecular mechanisms underlying canola salt stress tolerance.
Plant material and salt treatments
Seeds of the B. napus var. Global were germinated in a Metro-Mix 500 potting mixture (The Scotts Co., Marysville, OH, USA), and plants were grown in a growth chamber under a photosynthetic flux of 160 μmol of photons m-2 s-1 with a photoperiod of 10 h at 24°C in light and 12 h at 20°C in dark. When the seedlings reached four-week old, they were transplanted into pots filled with perlite and vermiculite (1:1), and watered with a half-strength Hoagland solution. Two weeks later, the seedlings were treated with 0, 50 mM, 100 mM, 200 mM and 400 mM NaCl with the salt concentration increased in aliquots of 100 mM every day until the 200 mM and 400 mM were reached. On day 3, 5, 7, 10, 12 and 14 after treatment, the third full expanded leaf from the top was selected for physiological assays or immediately frozen in liquid nitrogen and stored at -80°C for proteomics experiments. Three independent biological replicates for each control or treatment were conducted for all the experiments.
Physiological analyses of control and salt-stressed plants
Relative water content (RWC) of leaves: After different periods of salt treatment, fresh weight (FW) was measured immediately after the leaves were harvested. Dry Weight (DW) was obtained after drying the samples at 75°C for 48 hours. Turgor weight was determined by subjecting the leaves to rehydration for two hours after their fresh weight was determined. For each experiment, RWC was determined according to a previous method [58]: RWC(%)=(FW-DW) / (turgor weight-DW) × 100.
Relative Electrical Conductance (REC) of leaves: The REC of leaves was measured as described by Dionisio-Sese and Tobita 1998 [59], using a Fisher Scientific Accumet Excel XL30 conductivity meter. The REC was expressed as EL(%) = (EC1-EC0) / (EC2-EC0) × 100. EC1 represents the initial electrical conductivity of the medium, and EC2 indicates the final electrical conductivity of the medium after being autoclaved at 121°C for 20 min, while EC0 shows the electrical conductivity of distilled water.
Photosynthesis and chlorophyll fluorescence analysis: Gas exchange measurements, such as stomatal conductance (Cond), intercellular CO2 concentration (Ci), transpiration rate (Tr), and photosynthesis rate (Pn), were determined at 10:00 am with a LI-6400XT photosynthesis system (LI-COR Inc., Lincoln, NE, USA), and the respiration rate was determined at 10:00 pm in dark. Water-use efficiency (WUE) was calculated from Pn divided by Tr [8]. The chlorophyll fluorescence parameters (Fv/Fm and Fv/Fo) was measured using a Handy PEA portable fluorescence spectrometer (Hansatech Instruments, Ltd., King’s Lynn, UK) [60].
Antioxidant enzyme activity assay: Leaves of 0.5 g FW were homogenized in 100 mM potassium phosphate buffer (pH 7.8) containing 0.1 mM EDTA, 1 % (w/v) PVP, 0.5 % (v/v) Triton X-100, 5 mM ascorbate. The homogenate was centrifuged at 10 000 g for 20 min at 4°C. The supernatant was collected for measurements of antioxidant enzyme activities: 1) Superoxide dismutase (SOD) activity was determined as described by Stewart and Bewley [61]. The reaction mixture (3 ml) consisted of 50 mM phosphate buffer (pH 7.8), 13 mM methionine, 75 mM nitroblue tetrazolium, 100 mM EDTA and 2 mM riboflavin. After adding 0.1 ml of enzyme extract, test tubes were shaken and placed 30 cm below a light source (30 W fluorescent lamps). The reaction was started by switching-on the light. The reaction was allowed for 30 min and then stopped by switching-off the light. The tubes were kept in the dark until absorbance reading. The reaction mixture, which was not exposed to light did not develop color and served as control. The absorbance was measured at 560 nm in a spectrophotometer. Log A560 was plotted as a function of the volume of enzyme extract used in the reaction mixture. The volume of the enzyme extract corresponding to 50% inhibition of the reaction was read and considered as one enzyme unit and expressed as unit mg–1 protein. 2) Ascorbate peroxidase (APX) activity was determined in 3 ml of reaction mixture containing 50 mM potassium phosphate (pH 7.0), 0.5 mM ascorbate (extinction coefficient ε=2.8 mM cm–1), 0.1 mM EDTA, 1.5 mM H2O2 and 0.1 ml of the enzyme extract. The linear decrease in absorbance at the wavelength of 290 nm was followed for 1 min as described by Nakano and Asada [62]. The reaction was initiated by addition of H2O2. The activity of APX was calculated in terms of μmol ascorbic acid oxidized min–1 mg–1 protein. 3) Glutathione S-Transferase (GST) activity was measured at 25°C in a reaction mixture of 3 ml (1.5 ml 50 mM pH 7.0 phosphatic buffer, 0.3 ml 1 mM GSH, 0.3 ml 1mM 1-chloro-2,4-dinitrobenzene (CDNB), and 0.1 ml enzyme extract, and the reaction mixture without CDNB was used as control. The enzyme activity was calculated based on the change of absorbance at 340 nm in 1 min [63]. 4) Catalase (CAT) activity was determined by a modification of the spectrophotometric method [64]. The reaction mixture contained 50 mM phosphatic buffer (pH 7.0), 20 mM H2O2 and 0.1 ml enzyme extract. The reaction was initiated by addition of H2O2. The change in absorbance at 240 nm was monitored for 3 min. The enzyme activity was calculated based on the change of absorbance at 240 nm in 3 min (OD240•min-1•mg-1 protein).
Biochemical analysis of control and salt stressed plants
Analysis of soluble sugar, proline and betaine content: After sampling, fresh leaves were lyophilized and ground into fine powder for proline and sugar analysis. Total soluble sugar and proline contents were determined using ninhydrin reaction and an anthrone reagent, respectively [65]. Betaine content was determined with fresh leaves using Reinecke salt as previously described [66].
Statistical analysis of physiological and biochemical data: To determine whether there are significant differences in the data across different time points of salt stress treatment, analysis of variance (ANOVA) was performed using JMP 10.0.2 (SAS Institute, Cary, NC, USA). A two-sample t-test was performed to assess whether control and salt-stressed samples differ significantly at each time point.
Protein extraction, digestion, iTRAQ labeling and LCMS/ MS
Proteins were extracted and quantified as previously described [67], and dissolved in 0.1% SDS, 0.5 M triethylammonium bicarbonate, pH 8.5. For each sample, a total of 100 μg of protein were reduced, alkylated, trypsin-digested, and labeled according to the manufacturer’s instructions (AB Sciex Inc., Foster City, CA, USA). The control, 100 mM, 200 mM and 400 mM NaCl treated samples at 7 day were labeled with iTRAQ tags 113, 114, 115 and 116, and the corresponding 14 day samples were labeled with iTRAQ tags 117, 118, 119 and 121, respectively. Two independent experiments were carried out with different biological samples. Labeled peptides were desalted with C18-solid phase extraction and dissolved in strong cation exchange (SCX) solvent A (25% (v/v) acetonitrile, 10 mM ammonium formate, and 0.1% (v/v) formic acid, pH 2.8). The peptides were fractionated using an Agilent HPLC 1260 with a polysulfoethyl A column (2.1 × 100 mm, 5 µm, 300 Å; PolyLC, Columbia, MD, USA). Peptides were eluted with a linear gradient of 0–20% solvent B (25% (v/v) acetonitrile and 500 mM ammonium formate, pH (6.8) over 50 min followed by ramping up to 100% solvent B in 5 min. The absorbance at 280 nm was monitored and a total of 16 fractions were collected. The fractions were lyophilized and resuspended in LC solvent A (0.1% formic acid in 97% water, 3% acetonitrile). A hybrid quadrupole Orbitrap (Q Exactive) MS system (Thermo Fisher Scientific, Bremen, Germany) was used with high energy collision dissociation (HCD) in each MS and MS/MS cycle as previously described [68]. It interfaced with an automated EasynLC 1000 system (Thermo Fisher Scientific, Bremen, Germany). Each sample fraction was loaded and carried out a Acclaim Pepmap 100 pre-column (20 mm × 75 μm; 3 μm-C18) and an PepMap RSLC analytical column (250 mm × 75 μm; 2 μm-C18) with a flow rate at 300 nl/min of solvent A (0.1% formic acid) using 25% solvent B (0.1% formic acid, 99.9% acetonitrile) for 95 min, 98% B for 100 min.
Proteomics data analysis
The MS/MS data were processed by a thorough search considering biological modifications and amino acid substitution against a non-redundant Brassica database with decoy sequences (389,401 entries) using ProteoIQ v2.7 (Premier Biosoft, Palo Alto, CA, USA) and Proteome Discoverer v1.4 (Thermo Fisher Scientific, Bremen, Germany) with SEQUEST algorithm [69] and the following parameters: peptide tolerance at 10 ppm, tandem MS tolerance at ± 0.01 Da, peptide charges of 2+ to 4+, trypsin as the enzyme, allowing one missed cleavage, iTRAQ label and methyl methanethiosulfonate (C) as fixed modifications, and oxidation (M) and phosphorylation (S, T, Y) as variable modifications. Peptide and protein were filtered using ProteoIQ 2.7 with strict peptide and protein probabilities, 0.99 and 0.95, respectively. Peptide probability is applied to filter peptide assignments obtained from MS/MS database searching results using predictable false identification error rate [70]. Protein probability is used for filtering proteins with the likelihood that the protein assignment is correct taking into account of the peptide probability for all peptides apportioned to that protein [71]. For protein quantification, only MS/MS spectra that were unique to a particular protein and where the sum of the signal-to-noise ratios for the entire peak pairs >9 were used for quantification. The accuracy of each protein ratio is given by a calculated error factor, and a p value is given to assess whether the protein is significantly differentially expressed. The error factor is calculated as the 95% confidence error which is the weighted standard deviation of the weighted average of log ratios multiplied by Student’s t test. To be identified as being significantly differentially expressed, a protein should be quantified with at least three spectra (allowing generation of a p value), a p value <0.05, and a fold change >1.2 or <0.8 with at least six unique peptides in the experimental replicates (Supplementary Table 1). In addition, Single Enrichment Analysis (SEA) was performed with the list of differential proteins using agriGO (http://bioinfo.cau.edu.cn/ agriGO/) to determine specifically enriched biological processes. Proteins differentially expressed were clustered by hierarchical clustering via average linkage of Pearson correlations and the k-means clustering algorithm (k=8) using CLUSTER 3.0, and the results were visualized using Java TreeView (http://www.eisenlab. org). Subcellular location of the identified proteins was predicted using five internet tools: (1) YLoc (http://abi.inf.uni-tuebingen. de/Services/YLoc/webloc.cgi), with confidence score ≥0.4; (2) LocTree3 (https://rostlab.org/services/loctree3/), with expected accuracy ≥80%; (3) ngLOC (http://genome.unmc.edu/ngLOC/ index.html), with probability ≥80%; (4) TargetP (http://www. cbs. dtu.dk/services/TargetP/), with reliability class ≤3; (5) PlantmPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/), with no threshold value in Plant-mPLoc. Only the consistent predictions from at least two tools were accepted as a confident result. For the inconsistent prediction results among the five tools, subcellular localizations of corresponding proteins were obtained from literature if available.
Morphological changes of B. napus in response to salt stress
To determine how salt stress affects B. napus seedlings, we monitored their morphological responses under different salt concentrations treatment (0, 50 mM, 100 mM, 200 mM and 400 mM NaCl) at seven different time points (0, 3, 5, 7, 10, 12 and 14 days) (Supplementary Figure 1). Compared with the control plants, within 7 days of treatment under 50 mM, 100 mM, 200 mM or 400 mM NaCl, the leaves of salt-treated seedlings did not exhibit any obvious phenotype differences. After 10 days of salt treatment, when the salt concentration exceeded 100 mM, plant growth began to show slight inhibition. Some fully expanded leaves appeared chlorotic after 7 days of 400 mM NaCl treatment. After 14 days of salt stress and with increasing salt concentrations from 100 mM to 400 mM, plant growth was severely inhibited (Figure 1A). Although the seedlings could survive the 14-day 400 mM NaCl treatment, they began to wilt and lose viability.
Physiological and biochemical changes of B. napus in response to salt stress
The relative water contents (RWC) in leaves of B. napus seedlings were significantly lower in 200 mM and 400 mM NaCl treated plants than control plants from the 10th day to the 14th day of stress (Figure 1B). There was no significant difference between control and the 50 mM or 100 mM NaCl treated plants. The relative electrical conductivity (REC) in the leaves was significantly higher than control from the 3rd day of high concentration salt stress, indicating cell membrane damage by the salt stress (Figure 1C).
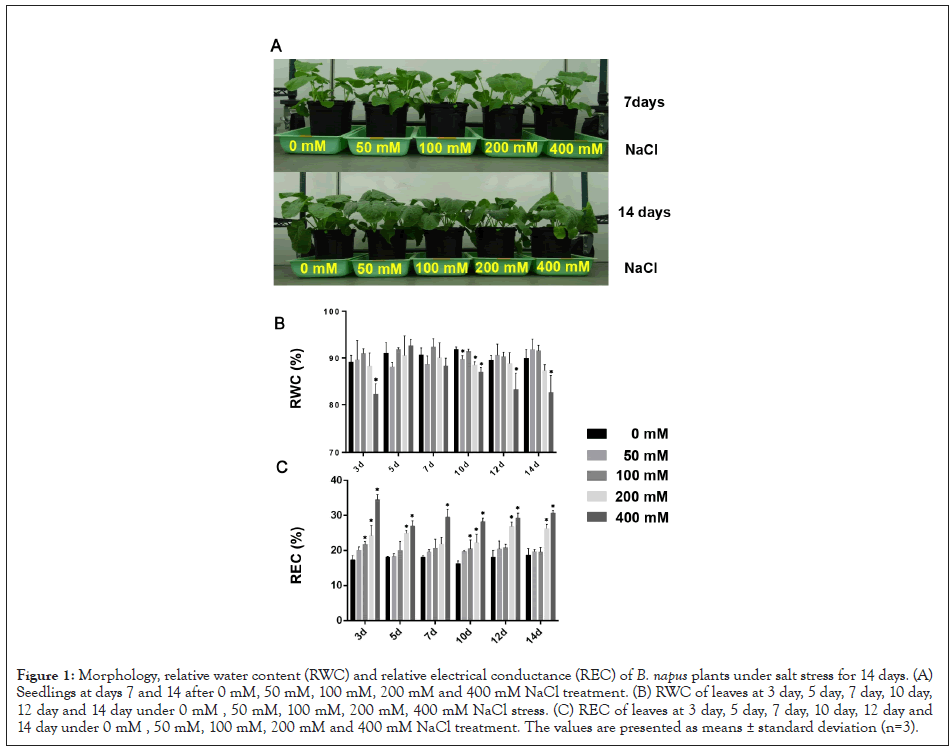
Figure 1: Morphology, relative water content (RWC) and relative electrical conductance (REC) of B. napus plants under salt stress for 14 days. (A) Seedlings at days 7 and 14 after 0 mM, 50 mM, 100 mM, 200 mM and 400 mM NaCl treatment. (B) RWC of leaves at 3 day, 5 day, 7 day, 10 day, 12 day and 14 day under 0 mM , 50 mM, 100 mM, 200 mM, 400 mM NaCl stress. (C) REC of leaves at 3 day, 5 day, 7 day, 10 day, 12 day and 14 day under 0 mM , 50 mM, 100 mM, 200 mM and 400 mM NaCl treatment. The values are presented as means ± standard deviation (n=3).
The overall photosynthetic efficiency including stomatal conductance (Cond), intercellular CO2 concentration (Ci), transpiration rate (Tr), photosynthetic rate (Pn), and the chlorophyll fluorescence parameters (Fv/Fm and Fv/Fo) decreased significantly at the 5th day of 200 mM and 400 mM salt stress treatment compared to the control samples (Figure 2A-2D, 2G, 2H). Interestingly, water use efficiency (WUE) was obviously increased after the 5th day of salt stress (Figure 2E). Although the respiration rate did not decrease in the beginning of the salt treatment, it showed remarkable decreases from the 10th and 12th day of 200 mM and 400 mM salt stress (Figure 2F).
Figure 2: Temporal photosynthesis and chlorophyll fluorescence analyses of B. napus plants under salt stress conditions. (A) Stomatal conductance (Cond); (B) Intercellular CO2 concentration (Ci); (C) Transpiration rate (Tr); (D) Photosynthesis rate (Pn); (E) Water usage efficiency (WUE); (F) Respiration rate; (G) Chlorophyll fluorescence parameter (Fv/Fm); (H) Chlorophyll fluorescence parameter (Fv/Fo). The data were collected at day 0, day 3, day 5, day 7, day 10, day 12 and day 14 under 0 mM , 50 mM, 100 mM, 200 mM and 400 mM NaCl conditions. The values are presented as means ± standard deviation (n=3).
The activities of ROS scavenging enzymes, such as SOD and APX increased in response to the 200 mM and 400 mM NaCl from the 7th to 14th day (Figure 3A, 3B), while the CAT activity decreased in response to the 200 mM and 400 mM salt stress at the 10th and 14th day (Figure 3C). The GST activities did not have apparent differences except an increase at the 7th day of 400 mM NaCl treatment compared to the control samples (Figure 3D). As to the biochemical responses, B. napus seedlings showed accumulation of stress metabolites such as betaine, proline and soluble sugar. Betaine levels increased at the 5th day after stress and showed drastic accumulation from the 7th to 14th day under the 200 mM and 400 mM NaCl (Figure 3E). Proline levels showed obvious increases from the 3rd day, and showed drastic accumulation from the 5th to 14th day (Figure 3F). In contrast, the soluble sugar level did not show remarkable differences after 200 and 400 mM salt stress (Figure 3G).
Figure 3: Antioxidant enzyme activity assay and contents of soluble sugar, proline and betaine in B. napus plants under salt stress. Superoxide dismutase (SOD) activity (B) Ascorbate peroxidase (APX) activity; (C) Catalase (CAT) activity; (D) Glutathione S-Transferase (GST) activity; (E) Betaine content; (F) Proline content; (G) Soluble sugar content. The data were collected at day 3, day 5, day 7, day 10, day 12 (for only E to G) and day 14 under 0 mM , 50 mM, 100 mM, 200 mM and 400 mM NaCl conditions. The values are presented as means ± standard deviation (n=3).
Identification of differentially expressed proteins in response to salt stress
The changing trends from physiological and biological analyses were not completely the same in the course of treatments with different concentrations of salt. For example, 50 mM NaCl treatment had no effect on B. napus seedlings within 14 days, while after 7 days of salt treatment, the seedlings began to be affected by salt stress, and after 14 days of salt treatment they were obviously affected and inhibited by salt stress. In order to investigate the salt tolerance mechanisms, we chose three salt concentrations (100 mM, 200 mM and 400 mM NaCl) and two time points (7 day and 14 day) based on their phenotypic and physiologica responses. Using iTRAQ and 2D LC-MS/MS, a total of 2316 proteins were identified in control and salt stressed samples at a 95% confidence level (Supplementary Table 1). The identified proteins covered a wide range of biological processes (Supplementary Figure 2).
Proteomic changes were examined between the control and 100 mM, 200 mM and 400 mM NaCl treated plants at the 7th day and 14th day of treatment. A total of 614 proteins were determined to be differentially expressed in salt stressed samples compared to control samples (a fold change>1.2 or <0.8, p<0.05) (Supplementary Table 2). The differential proteins were grouped into ten functional categories based on the biological processes according to the published literatures and UniProt online analysis (https://www.uniprot.org/) (Figure 4A). They are involved in metabolism (21%), stress and defense (19%), photosynthesis (16%), protein folding and degradation (11%), transport (9%), protein synthesis (8%), signal transduction and kinase (7%), cell structure (4%), transcription related (4%) and unknown (1%). Based on agriGO functional enrichment analysis of the 614 DEPs, the DEPs involved in cellular, metabolic and response to stimulus processes were enriched under salt stress (Supplementary Figure 3). Subcellular localization analysis of the 614 DEPs revealed the top four subcellular locations as chloroplast, cytoplasm, mitochondria and nucleus (Figure 4B).
Figure 4: Functional categorization and subcellular localization of the 614 salt-stress responsive proteins in leaves of B. napus plants (A) The saltstress responsive proteins were classified into 10 functional categories. The percentage of proteins in each functional category is shown in the pie (B) Subcellular localization groups of the 614 proteins. The percentage of proteins in each localization is shown in the pie.
Expression patterns of the 614 DEPs fell into 27 categories (Supplementary Table 3). At 7 day of salt stress, there were 77 increased and 54 decreased proteins under 100 mM NaCl treatment (Supplementary Table 4), 171 increased and 35 decreased proteins under 200 mM NaCl treatment (Supplementary Table 4), and 233 increased and 99 decreased proteins under 400 mM NaCl treatment (Supplementary Table 4). At 14 day of salt stress, there were 396 increased and 29 decreased proteins under 100 mM NaCl treatment (Supplementary Table 5), 114 increased and 19 decreased proteins under 200 mM NaCl treatment (Supplementary Table 5), and 170 increased and 14 decreased proteins under 400 mM NaCl treatment (Supplementary Table 5).
Salinity-responsive protein-protein interactions
Proteins rarely act alone, and knowledge of the protein-protein interactions (PPIs) will facilitate molecular studies on diverse biological processes and provide insight into the biological functions of proteins with unknown functions [72]. The Biological General Repository for Interaction Datasets (BioGRID: http// thebiogrid.org) is an open access archive of genetic and protein interactions that are curated for all major model organism species [73]. To discover the relationship of the 614 differentially expressed proteins in response to salt stress, PPI networks were generated using the BioGRID database. After BLASTing in TAIR database (http://www.arabidopsis.org/Blast/index.jsp), 605 homologs in Arabidopsis of the 614 differential proteins were analyzed, and then subjected to the molecular Interaction BioGRID database for creation of proteome-scale interaction network. Among them, 138 proteins were depicted in the BioGRID database, and illuminated in six functional modules with tightly-connected clusters in the network (Figure 5 and Supplementary Tables 2 and 6). A total of 61 proteins were connected in the red module. There are six 14-3-3 proteins in the red module, and these 14-3-3 proteins are at the core, indicating that 14-3-3 proteins interact with multiple proteins to participate in signal transduction. In addition, 49 proteins were connected in the green module with the 26S proteasome at the core, indicating that it interacts with multiple proteins for potential degradation.
Figure 5: Protein–protein interaction (PPI) network in B. napus plants revealed by BioGRID analysis. A total of 138 salt responsive proteins represented from the corresponding Arabidopsis proteins are shown in PPI network. Six main groups are indicated in different colors. The PPI network is shown in the confidence view generated by BioGRID database. The abbreviations refer to Supplementary Tables 2 and 6.
Salt stress affects plants at different levels, including physiological, biochemical and molecular processes, e.g., ionic imbalance, hyperosmotic stress, oxidative damage and nutrient deficiency [21,23,74]. Here we report salt stress responses in B. napus seedlings using an integrated physiological, biochemical, and proteomic approach.
Increased photosynthetic proteins to compensate for the decline of photosynthetic efficiency in B. napus leaves under salt stress
In this study, 18 proteins associated with photosynthesis were identified, 16 of which were increased and they were oxygenevolving enhancer protein 2 (OEE2), OEE3, oxygen-evolving complex (OEC), chlorophyll a b-binding protein (CAB) , cytochrome b6-f complex protein (Cytb6f), ferredoxin NADP+ reductase (FNR), Ribulose 1,5-biphosphate carboxylase/oxygenase (RubisCO), RubisCO activase (RCA), phosphoglycerate kinase (PGK), glyceraldehyde-3-phosphate dehydrogenase (GADPH), triosephosphate isomerase (TPI), transketolase (TK), ribulose 5-phosphate isomerase (RPI), phosphoribulokinase precursor (PRK), magnesium-chelatase subunit chli and protochlorophyllide reductase (POR). Only a carbonic anhydrase (CA) was decreased. These proteins are involved in light reaction, Calvin cycle and chlorophyll synthesis pathways in photosynthesis. The expression levels of several proteins under salt stress are consistent with previous results. For example, OEE 2 and OEE3-1 were increased in soybean, Vigna unguiculata and barley under salt stress [75-77]. The CABs were also significantly increased in stress proteomics of mulberry and tea [78,79]. Cytb6f was steadily increased, and this protein was recognized as one of the salt stress-responsive markers in wheat chloroplasts [80]. The early reported increase in TK in Arabidopsis also agreed with the changes in TK activity observed in Z. mays under salt and oxidative stresses [79] and supported the hypothesis that TK was involved in plant stress protection. Proteomic studies of celery leaves revealed that magnesium chelatase (Mg chelatase) related to Chl biosynthesis showed increased abundances in cold-stress [81]. However, not all the protein changes under salt stress are consistent with previous studies. For example, a 33 kDa protein in the photosystem II oxygen-evolving complex (OEC) in white clover was decreased in abundance late in senescence, indicating degradation of photosynthetic apparatus during leaf senescence [82]. In this study, the increased OEC level showed that the photosynthetic apparatus was still stable for B. napus plants under salt stress for 14 days. In another proteomic study upland cotton leaves, PORs were markedly suppressed after 200 mM salt treatment for 4 h, 8 h, 24 h, 48 h and 72 h) [83,84]. Clearly, B. napus plants may use different mechanisms to adapt to salt stress conditions (see below).
Physiological changes of B. napus in response to salt stress showed the parameters related to photosynthetic efficiency (Figure 2) showed remarkable decreases compared to the control samples after the 5th day of 200 mM and 400 mM salt stress. The results are consistent with previous reports in that salt stress provoked rapid stomatal closure and impairment in CO2 assimilation, leading to significant decreases in photosynthetic efficiency [3]. The decrease of photosynthetic efficiency may be related to cellular oxidative state under salt stress. For example, a glycolate oxidase (GO) in peroxisomes increased and can oxidize glycolate from chloroplasts to glyoxylate [85], leading intracellular H2O2 accumulation. Under salt stress, the activities of antioxidant enzymes (e.g., SOD and APX) increased (Figure 3). This is generally a response to increased ROS and oxidative stress [85,86]. ROS scavenging enzymes (i.e., SOD, CAT, Prx, Trx, APX, DHAR, MDHAR, GPX, GR and GST) were overrepresented in DEPs under salt stress, especially at 14 day after stress (Figure 6C). Cellular ROS scavenging system includes catalase metabolic pathway, peroxiredoxin-thioredoxin pathway (PrxR-Trx cycle) and ascorbate-glutathione cycle (AsA-GSH cycle) [86]. Under salt stress, B. napus cells enhanced the ROS scavenging activities to avoid oxidative damage. Nonetheless, the cells may be still under an oxidative state, as evidenced by the increased of three protein disulfide-isomerase (PDI) under salt stress (Figure 6F). PDIs are multifunctional enzymes able to facilitate oxidative folding of nascent secretory proteins in the endoplasmic reticulum (ER) [87,88]. Now how to explain the contradiction between the decreased photosynthetic efficiency and increased protein expression levels in photosynthesis in B. napus under salt stress? Under oxidative state, the proteins involved in photosynthesis tend to be inactive. As a positive feedback response, B. napus plants increased the expression of photosynthesis-related proteins (Figure 6A) as enabled by the upregulated expression of translational machinery (e.g., ribosomal proteins in Supplementary Table 2). This response compensates for the decrease of photosynthetic efficiency in B. napus, allowing the plants to maintain photosynthesis and acquire tolerance under salt stress.
Figure 6: Schematic presentation of salt tolerance mechanisms in B. napus plants (A) Proteins in photosynthesis pathway. The proteins were integrated into light reaction, chlorophyll synthesis and Calvin cycle; (B) Proteins in glycolysis and the citric acid cycle; (C) Proteins involved in ROS scavenging and glyoxalase systems. The two arrows from left to right indicate days 7 and 14, respectively, with red and green showing increased and decreased, respectively. The blue arc and black lines indicate salt-stress responsive proteins and unresponsive proteins, respectively; (D) Proteins involved in signal transduction and kinases; (E) Proteins involved in iron transport; (F) Proteins involved in protein folding and degradation. Protein expression patterns under salt stress were shown by marking the proteins and reactions in red for increased proteins and in dark green for decreased proteins. Please refer to text and Supplementary Tables 2 and 7 for abbreviations.
Detoxification of methylglyoxal (MG) by the glyoxalase system in B. napus
Under salt stress, the decreased stomatal conductance, leaf intercellular CO2 levels and CA levels (Figure 2A, 2B and 6A), together with oxidative stress caused the decrease of photosynthesis [89]. Therefore, less oxygen was released from light reaction and the respiration rate was decreased, resulting in anaerobic respiration for limited energy production. In spite of significant increases of key enzymes involved in glycolysis (Figure 6B), the citric acid cycle of aerobic respiration was not functional. Under salt stress, glycolysis pathway could produce a highly reactive dicarbonyl metabolite, methylglyoxal (MG), through degradation of triose phosphates, glyceraldehyde-3- phosphate, and dihydroxyacetone phosphate [90]. MG has a dual role in plant cells, as a cytotoxin at high concentration or a signal molecule at low concentration [91,92]. Thus, MG homeostasis in plant cells is critical. When MG levels increase under different abiotic stresses, the glyoxalase system plays an important role in the detoxification of MG. The glyoxalase system (the glyoxalase pathway) is composed of two enzymes glyoxalase I (Gly I) and glyoxalase II (Gly II). Gly I detoxifies MG to S-lactoylglutathione (SLG) by using one molecule of reduced glutathione (GSH) [91,93]. Subsequently, SLG is converted to lactose by Gly II and one molecule of GSH is recycled back into the system [91]. In this study, five glyoxalases I (Gly I) and one glyoxalase II (Gly I) were increased in salt stressed B. napus plants for 7 and 14 days (Figure 6C).
In consistent with our results, overexpression of Gly I or Gly II genes in rice, Carrizo citrange, tomato and sugar beet plants conferred transgenic plants tolerance to salt stress and other abiotic stresses [94-97]. Transgenic plants were able to survive the various abiotic stresses through maintaining cellular MG and GSH homeostasis. Recently, the expression patterns of 16 BrGLY I and 15 BrGLY II genes from Chinese cabbage (B. rapa) were analyzed in different tissues and their responses to biotic and abiotic stresses. A number of BrGLY genes appeared highly responsive to these stress treatments, including salt stress, Plasmodiophora brassicae infection and heavy metal stress [98].
Increased signal transduction and ion transport activities to enhance salt tolerance
In plants, 14-3-3 proteins are a family of highly conserved proteins that not only regulate a variety of biological functions including signal transduction, but also interact with proteins phosphorylated on Ser or Thr residues in a conserved binding motif [99-101]. In this study, nine 14-3-3 and 14-3-3-like proteins were all increased under salt stress for 7 and 14 days (Figure 6D). Only one 14-3-3-like protein was decreased under 400 mM NaCl for 7 days. In salinity-responsive protein-protein interactions, there are six 14-3-3 proteins at the core in the red module (Figure 5). In general, the 14-3-3 interaction can change target protein localization, conformation, stability, activity, or affinity to other proteins [102,103]. 14-3-3 proteins have been reported to function in biotic and abiotic stress responses, such as salt stress [104,105]. For example, ectopic expression of a 14-3-3 adaptor gene TaGF14b from wheat enhanced drought and salt tolerance in transgenic tobacco [106]. TaGF14b expression led to enhanced ROS scavenging to ameliorate oxidative damage. In addition, a 14-3-3 gene in Brachypodium distachyon, BdGF14d, conferred salt tolerance in transgenic tobacco plants through regulating ABA signaling, ROS-scavenging, and ion transport [107].
Consistent with our results, proteomics of leaves of two upland cotton genotypes differing in salt tolerance showed that a 14-3- 3-like protein E was induced by salt stress [84]. Proteomics and phosphoproteomics of sugar beet monosomic addition line M14 under salt stress showed that although the 14-3-3 protein levels did not change between salt stress and control samples, its phosphorylation modification changed under 200 mM NaCl [108]. Four 14-3-3 proteins in soybean leaves and three 14-3-3 proteins in soybean roots were dramatically increased at transcriptional and translational levels [109]. Salt-Overly- Sensitive (SOS)2-LIKE PROTEIN KINASE5 (PKS5) negatively regulates the SOS signaling pathway in Arabidopsis [110]. Under normal growth conditions, PKS5 phosphorylates SOS2Ser294 and represses SOS2 activity. Upon salt stress, 14-3-3 proteins decode a salt-induced calcium signal, repress PKS5, and release SOS2 to activate H+-ATPase and SOS1(Na+/H+ antiporter), thus increasing Na+ efflux [110]. This mechanism may work similarly in B. napus salt-tolerance.
In addition to 14-3-3 proteins, four histidine kinases (HK) and three two-component system response and regulator (RR) were increased under salt stress (Figure 6D). Two component systems are found in bacteria, archaea, fungi, slime molds, and plants [111-113]. The receptor HK autophosphorylates on a conserved histidine residue in response to salt stress, and the phosphate is then transferred to a conserved aspartic acid residue within the receiver domain of RR proteins [114]. RR proteins frequently function as transcription factors, and phosphorylation modulates their regulation of gene expression [114]. The two-component signaling system has an established role in mediating cytokinin and ethylene signal transduction in plants [114,115]. Here the two-component signal transduction system may play an important role in transmitting salt stress signals, regulating the expression of downstream salt responsive genes to initiate salt stress response and tolerance.
In this study, six V-type H+-ATPase (VHA) and five Plasma membrane ATPase (PMA) were all increased (Figure 6E). PMA, VHA and pyrophosphatase (PPase) are major proton pumps, providing energy for ion transport across plasma membrane and tonoplast, respectively [116,117]. Under high salt conditions, plants use an electrochemical H+-gradient generated by the H+ pumps to activate secondary transport of Na+ from the cytosol into the vacuole or to extracellular space [118,119]. When Arabidopsis VHA-c1 and c3 subunit isoforms were knocked down by RNAi, each resulted in reduced root length and decreased salt stress tolerance [120]. This result is consistent with the salt stress induced expression of VHA genes and VHA proteins in mature sugar beet leaves [121]. Ectopic expression of a wheat VHA gene in Arabidopsis and an Arabidopsis VHA gene in barley both improved salt tolerance of transgenic plants [122,123]. Therefore, the induced H+ pumps in B. napus plants under salt stress helped to decrease cytosolic Na+ levels, eliminate Na+ toxicity and enhance salt resistance.
Increased proteins involved in protein folding and degradation under salt stress
Many proteins involved in protein folding and degradation were increased in B. napus plants under salt stress, including 12 heat shock proteins (HSPs), 12 chaperone/chaperonin proteins, 9 peptidyl-prolyl cis-trans isomerase cyclophilins chaperone proteins (CYP), 10 proteasome proteins, and 3 protein disulfideisomerase (PDI) (Figure 6F). Consistent with our results, HSP70 expression was enhanced in salt-treated sugarcane [124], and in salt-stress tolerant potato plants [125]. Overexpression of HSP70 in A. thaliana led to decreased membrane damage and remarkable tolerance to heat, drought and salinity compared to the wild type control plants [124-128]. These studies concluded that HSP70 is crucial to alleviate the damage of membrane peroxidation, especially in chloroplasts, and thus confers salt tolerance.
CYPs are ubiquitous proteins found in bacteria, fungi, insects, plants and mammals. Most of them, if not all, have peptidylprolyl cis-trans isomerase (PPIase) activities [129]. CYP expression has been shown to be induced by biotic and abiotic stresses [130-133]. A hypothetical model depicting possible mechanisms through which cyclophilins exert their stress protective properties (Figure 6F) includes ROS Scavenging, folding of nascent proteins, refolding of aggregated proteins, posttranscriptional gene silencing, cellular protection and DNA repair [129]. Proteasomes constitute one of the main cellular proteolytic mechanisms. They recognize and degrade peptides and proteins to maintain the equilibrium between normal protein production and degradation, or to eliminate the damaged, misfolded/unfolded or pathogenic proteins [134]. The most important protease increased during the proteotoxic stress is the 26S proteasome [135,136]. This ATP-dependent proteolytic machinery works in tandem with ubiquitin to direct the selective breakdown of aberrant proteins and short-lived proteins [137]. Such capabilities are important for plant salt-stress tolerance.
Other metabolic and stress and defense proteins important for salt stress tolerance
Five cysteine synthases (CS) were all increased salt stress for 7 and 14 days (Figure 6B). CSs are key enzymes for synthesizing L-cysteine from L-serine. In consistent with our result, another proteomic study of salt stress responsive proteins in roots and leaves of rice showed that a CS was increased under salt stress [138]. In addition, a CS gene from Polygonum sibiricum Laxm (PcCSase1) was transformed into yeast cells. The survival rate of the transgenic yeast was higher than the control under 10% NaHCO3 and 5 M NaCl stress. The results proved that PcCSase1 confers high salt tolerance in yeast cells by increasing the contents of cysteine and glutathione [139].
Three ATP sulfurylases (ATPS) were increased under salt stress for 7 and 14 days (Figure 6B). The ATPS mediates selenate (Se) reduction, and promotes Se and sulfur (S) uptake and assimilation. The roles of nitric oxide (NO) and sulfur (S) on stomatal responses and photosynthetic performance were studied in mustard (B. juncea L) under salt stress [140]. These plants receiving NO plus S exhibited increased activities of ATPS and antioxidant enzymes, minimizing oxidative stress. Four glutamine synthetases (GS) were also increased in salt stressed B. napus plants (Figure 6B). The GSs are involved in nitrogen metabolism via ammonium assimilation which catalyzes the ATP-dependent biosynthesis of glutamine from glutamate and ammonia [141]. Consistent with our results, the salt tolerance of A. thaliana and Z. mays was enhanced by inoculation with Bacillus amyloliquefaciens SQR9, which produces spermidine. Spermidine was shown to increase GS gene expression, leading to increased levels of GSH, which is critical for ROS scavenging [104].
Through morphological, physiological and biochemical analyses, together with high-coverage iTRAQ-based quantitative proteomics, we conducted a comprehensive temporal study of B. napus salt-stress response and tolerance under different salt-stress conditions. A total of 614 DEPs were identified to include a range of biological processes, including stress and defense, metabolism and photosynthesis. As shown in Figure 6, the salt tolerance strategies of B. napus mainly include: (1) Increased levels of proteins associated with photosynthesis is a feedback mechanism to compensate for the decline of photosynthetic efficiency under salt stress; (2) Anaerobic respiration caused by stomatal closure and decreased photosynthesis led to cytotoxic MG production. Enhanced activities of the glyoxalase system and antioxidant systems can facilitate MG detoxification and ROS scavenging, respectively, for cellular redox homeostasis; (3) Increased 14-3-3 proteins, HKs and two-component system RR proteins ensure efficient stress signal transduction and activation of proton pumps for ion balance; (4) Increased HSPs, CYPs, proteasomes and PDIs involved in protein folding and degradation under salt stress are important salt-stress adaptive responses. This study revealed interesting salt tolerance mechanisms in B. napus, which lay the foundation for functional studies and potential applications in molecular breeding of stress-tolerance crops.
Refer to Web version on PubMed Central for supplementary material.
Appendix S1. A PDF file of Supplementary Figure 1 to 3 and Supplementary Table 3, 6 and 7.
The proteomics data generated in this study have been submitted to Massive repository (MSV000084676) and can be accessed via username ybgirl, password 123456789 and proteomeXchange# PXD016710.
This research was supported by the National Science Foundation of China (Grant No. 31671751 and 31801426), the Natural Science Foundation of Heilongjiang Province (Grant No. C2017057), the Common College Science and Technology Innovation Team of Heilongjiang Province (Grant No. 2014TD004) and China Postdoctoral Science Foundation (204968).
Citation: Yu B, Chen G, DuanMu H, Dufresne D, Erickson JE, Koh J, et al. (2021) Physiological and Proteomic Analysis of Brassica napus in Response to Salt Stress. J Proteomics Bioinform. 14:523.
Received: 26-Jan-2021 Accepted: 09-Feb-2021 Published: 16-Feb-2021 , DOI: 10.35248/0974-276X.21.14.523
Copyright: © 2021 Yu B, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.