Drug Designing: Open Access
Open Access
ISSN: 2169-0138
ISSN: 2169-0138
Research Article - (2022)Volume 11, Issue 3
Design of a drug compound that can effectively bind to the inside surface of E channel and block the diffusion of H+/K+ ions through channel and inhibit virus replication is an important task. A set of drug candidates, derivatives of a lead compound (diazabicyclooctane), were proposed early and its interaction with native and mutant structures of E protein are investigated via MD simulations and binding position/energies simulations. It is shown that E protein has in-channel and out of channel binding sites of high affinity for a set of molecular blockers. The most probable mutants of amino acids of E channel are considered and effectiveness of in-channel binding of blocker molecules is analyzed.
SARS-CoV-2 ; E channel inhibitors; Molecular dynamics; Ligand binding
The development of an effective anti-covid19 drug is an important task because of the grate ability of SARS-CoV-2 virus to infect fast a large population. A reasonable approach to affect the functional cycle of the SARS-CoV-2 virus particle by a specific affection of a functionally important element of virus particle, for example, the protein forming the trans-membrane channel E, an important element of proper functioning of virus particle. The E channel is formed by amino acid residues 8-65 of each of the five subunits A, B, C, D, E and a set molecular blockers are proposed and preliminary investigated by us [1]. The five interacting α-helices build up the E channel, which has central channel of large diameter ~17-19 Å if measured between Cα atoms of the sixteen NMR structures, PDB code 5X29 solved at T=308K, pH=5.5, 50 mM NaCl [2]. The E channel activity of the SARS-CoV-2 virus as it was shown is critical for infection process of living cell. It was shown that virus activity was significantly lower when the transmembrane segment, i.e. E protein, of infectious bronchitis virus was replaced by a heterologous domain without of the E ion channel activity [3]. For SARS-CoV-1 and SARS-CoV-2 viruses have been shown that decreased the E channel activity lead to far less infectious ability [4-6], to low virus assembling, pathogenesis and release [7,8]. The proton transport activity of E proteins channel from several coronaviruses, including, SARS-CoV-1 [9,10], MERS coronavirus [11], mouse hepatitis virus [12] and infectious bronchitis [13], may be blocked by molecular “cork”, molecule of hexamethylene amiloride [13,14]. Approaches to specifically affect the ion channel M2 proton channel of influenza A virus is suggested [15,16]. Proton diffusion through M2 channel can be suppressedvia binding of a special blocker molecule inside the ion channel that physically prevents proton diffusion through the channel [17,18]. It is crucial that E proteins are important for viral pathogenesis [19-21], therefore binding of crock molecule inside the E channel may destroy its proper function. Right activity of E channel of SARS-CoV-2 coronavirus was shown to be critical for infection process of living cell [3]. Here, a set of drug molecules, derivatives of 1,4-diazabicyclo-[2.2.2]octane (DABCO) were suggested by us [1] and investigated its ability to bind to native and mutant E protein ion channel affect a proper functioning of E channel. Blocker molecules suggested are effectively interacted with the E channel of SARS-cov2 virus and should considerably affect a proper functioning of E channel and, in the hole, on virus life circle.
Method of modeling
Atomic structure and temperature dynamics of E protein, at human body temperature, were modeled by using the fifteen NMD PDB structures, (pdb code 5X29), which are representing thermal dynamics at temperature 308K and pH 5.5 [2].
Modeling described in the paper were done by three computational techniques, energy optimization and MD simulations of protein in an environment, and exhaustive docking of a drug molecules to protein E in a lipid bilayer. MD simulation of equilibrium thermal fluctuations of three-dimensional structure via BioPASED software [22,23] with implicit solvent model, corrected Generalized Born approximation GB-MSR6c-pK [24]. Exhaustive molecular docking of drug molecules to the E protein was done by the hbDOCK method [25,26]. Electrostatic methods FAMBE and FAMBE-pH have been used for calculation of ionization constants pKa and of the ionization degree as a function of solvent pH [26,27]. All atomatom interactions are calculated with AMBER94 and AMBERGAFF [27-30] force field.
The protein forming E channel, PDB code 5X29, is embedded into the lipid cell membrane. Experimental measurement indicates that dielectric constant D0 of the lipid membrane (i.e., outside E protein volume (30)) is equal to ~30. The E protein volume and lumen volume of the E ion channel itself are considered here as the internal protein volume with a dielectric constant Di of 12 [31-37].
Structure of E protein channel
The E protein is unique in a sense that it has only one ionizable residue GLU8A (B, C, D, F) in each of five equivalent α-helices with pKa0=4.25. Therefore, the experimental NMR structure of E protein at pH=5.5 has ionized residues GLU8A (B, C, D, E), so that the ionized state of these residues will be the same at physiological pH 6-8 units. Thermal dynamics of E protein, PDB 5X29, is presented by the 16 NMR models witch reflect conformational flexibility of E protein structure at temperature 308K, Figure showing average RMSD 3.4 Å. The most critical mutants of residue of the E ion channel are at the entrance to the E channel, namely, GLU-8A (B, C, D, E) and ASN15A (B, C, D, E) which have a large probability to be mutated and replaced by neutral GLN and charged ASP- residues, respectively. Such mutations will considerably affect electrostatic properties of E channel and ability to bind charged drug molecules and probability of diffusion of H+ and K+ ions through the E channel (Figure 1).

Figure 1: NMR structures of E protein channel, of SARS-CoV-2 virus, all atoms RMSD ~3.4 Å, T=308K, pH=5.5, 50 mM NaCl. Sixteen structures reflect thermal dynamics of E protein, PDB cod 5X29. Five equivalent protein chains A, B, C, D, E are highlighted by colors.
Novel inhibitors of E channel
The 3D structure of the E protein, PDB code 5X29, shows that ion channel consists of the pentamer of five equivalent α-helices, which form the molecular E channel, residues 8-38 of each of five protein chains. Segment of residues 39-65 forms the outer protein shell of the E channel [2]. Function of E channel consists of a pumping of H+ and K+ ions through the cell membrane, similar to the function of the M2 channel of influenza A virus [17]. Activity of the E channel can be destroyed via putting a molecular “cork” inside the E channel, i.e. a specially designed molecule, which is able to effective binding inside the E channel and sterically and electrostatically prevent transport of positively charge cation through the channel, as it is shown for M2 ion channel of influenza A virus [18]. It should be noticed that it is shown that the E channel activity can be inhibited by Gliclazide and Memantine molecules [5]. A new class of molecular inhibitors of the E channel is based on diazabicyclooctane (DABCO), N2 (C2H4)3 [1].
Polysyclic derivatives
Polycyclic derivatives of DABCO consist of a larger number of atoms, compare to DABCO itself, and will have large van der Waals interactions with inside surface of E channel. A set of molecular blockers considered here presents derivatives of leading molecular structure DABCO, and have been considered early [1]. Namely molecules diazabicyclooctan-benzene (DABCOB), and diazabicyclooctan-naphthalene (DABCON), diazabicyclooctan-3- benzene (DABCO3B) and diazabicyclooctan-3-toluene(DABCO3N) [1].
Rationale for selecting the chemical class as BRAF inhibitors
Heterocyclic compounds are widely distributed in nature and are essential to life in various ways. Nitrogen-containing aromatic heterocyclic rings such as benzimidazole, pyrazine, and pyridine are important structural units in natural and synthetic pharmaceutical compounds. These heterocyclic rings have shown various pharmacological activities. Moreover, studies have shown heterocyclic compounds containing nitrogen, oxygen, and sulphur atoms are the core structures of several biologically active compounds. Amongst all heterocyclic compounds, nitrogencontaining8 rings like benzimidazole, pyridine, piperidine, pyrazine, etc. are frequently employed citing their resemblance to nucleic acid bases [46,47]. This property is considered very useful when diseases with underlying gene mutations are to be treated. As mentioned above, gene mutations have been identified as one of the causes of cancer; just like BRAF mutation. In this case, a mutation in the V600E site of BRAF gene locks it in the active state. As a result, BRAF continuously keeps sending signals to MEK protein for cell proliferation. Phosphorylation of human protein kinases is an important mechanism for signal transduction, where adenosine triphosphate (ATP) acts as a phosphodonor. This indicates the importance of nitrogen-containing heterocyclic ring systems in disease therapy and explains the success of dabrafenib [48] and vemurafenib [49] as BRAF inhibitors as shown in Figure 1. Hence, it was decided to study the potential of some of the pyrazine and benzimidazole derivatives as B-RAF inhibitors (Figure 2).
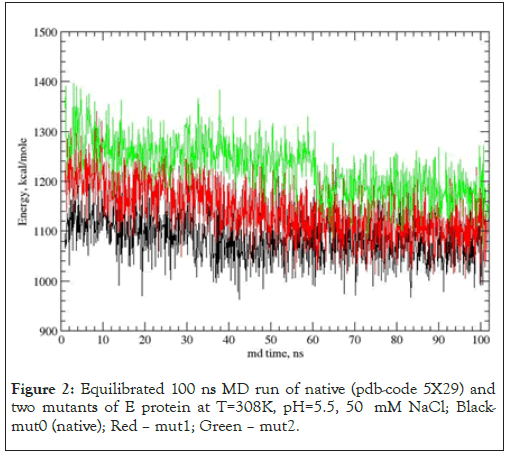
Figure 2: Equilibrated 100 ns MD run of native (pdb-code 5X29) and two mutants of E protein at T=308K, pH=5.5, 50 mM NaCl; Blackmut0 (native); Red – mut1; Green – mut2.
Native and mutant E protein channel
Preparation of low energy protein structure set for drug binding modeling was done via the next steps, a) fifteen NMR structures of PDB entree 5X29 were equilibrated in the force field user here, AMBER-GAFF [27-29] force field; b) the lowest energy initial NMR structure of protein E was taken for further modeling. Similarly, two mutant structures of E channel were prepared, so three structures of protein E are considered, namely, native, mut0, with GLU-8A(B, C, D, E) and ASN15A(B, C, D, E), mut1 with GLU-8A(B, C, D, E), ASP-15A(B, C, D, E), and mut2 with GLN8A(B, C, D, E) and ASN15A(B, C, D, E). These mutants highlighted role of acid, negatively charged residues at the entrance to the E channel. The 100 ns equilibrated MD trajectories of native and mutant forms of E channel are shown below Figure 2 and Table 1.
| aMutant | beProt | ceVDW | dCoul | eeHb | fDef | grel | heRMSD |
|---|---|---|---|---|---|---|---|
| native/ | 1075.5 | -1078.4 | -1441.5 | -1073.5 | 5194.1 | 0 | 53.2 |
| mut0 | |||||||
| mut1 | 1106.2 | -1082.1 | -1429.2 | -1042.8 | 5170.5 | 30.7 | 52.1 |
| mut2 | 1171.1 | -1074.1 | -1391.6 | -1066.7 | 5205.6 | 94.4 | 49.3 |
Note: aProtein structure; bTotal average energy of E protein mutant, kcal/mole; cEnergy of van der Waals interactions; dEnergy of columbic interactions; eH-bond energy; fEnergy of temperature deformation of molecular structure, gTotal energy relative to native structure; hEnergy of temperature fluctuations.
Table 1: Average energy of native and mutant structures of E protein.
Table 1 shows average values of five different types of total potential energy and different types of interactions for native and mutant structures along MD trajectories. It can be seen that native structure, mut0, with GLU-8A(B, C, D, E) and ASN15A(B, C, D, E) has a minimal potential energy along MD trajectory. Mutant mut1 with GLU-8A(B, C, D, E), ASP-15A(B, C, D, E), and mut2 with GLN8A(B, C, D, E) and ASN15A(B, C, D, E). Figure 2 shows that the native structure of E channel has the lowest average total potential energy over MD trajectory, while mutant structures have a large energy, mut1 by 30.7 and mut2 by 94.4 kcal/mole, respectively. The fluctuating potential energies of native, mut0, and mutants mut1 structures demonstrate a large degree of mutual overlapping in Figure 2. A strong overlap of potential energies along MD trajectories suggest that neutral residues ASN15A (B, C, D, E) can be spontaneously transformed into negatively charged acid ASP15A (B, C, D, E) with a large probability. Transition between neutral and charged states can be between different combinations of ionized/neutral, states of five residues, there by the new degree of freedom appeared, i.e. ionization state of five ASN/ASP residues appears, and respective entropy related term equal to –kTln(Niz) ~-kT*lnNiz, where Niz (=32) is the number of ionization states for five ASN15/ASP15(A, B, C, D, E) residues. A large overlap between energy fluctuations of native and mut1 structures suggests that spontaneous transitions between native and mut1 structures are the quite probable events. An energy overlapping between native and mut2 states is not significant, therefore a simultaneous appearance of neutral state of residues GLN8A (B, C, D, E) and ASN15A (B, C, D, E) is considerably less probable.
Binding of leading compound to native E protein channel
Molecular blockers investigated by us presented in Figure 1 are investigated on its binding with E protein, which forms the E ion channel of SARS-CoV-2 virus particle. Also, dependence of drug binding energy and binding position on the ionization state of GLU8 and ASN15 residues of the five chains of E protein are investigated. Exhaustive docking has been performed on a flexible structure of E channel of the SARS-CoV-2 virus, modeled by NMR structure, PDB code 5X29. Protein molecular surface accessible for water molecules was analyzed via the hierarchical blind docking method, HBDock, technique [25]. It must be noted that the HBDock method implements exhaustive docking of a flexible drug molecule to a flexible protein molecule via a multiple simulated annealing method and global optimization over rotational and translational degrees of freedom of a flexible drug molecule. A blocking of proton diffusion through the E channel has basically electrostatic nature due to the positive charge, +2, of the DABCO molecule. Steric blockage is negligible for proton H+ even it is solvated by six water molecules. It can be noted that the in-channel binding mode with energy of -24.3 kcal/mole, has the energy gap relative of out-of-channel binding mode, EGAP=EOUT — EIN ~0.5 kcal/mole, i.e. ratio of IN and OUT channel bound fractions is noticeable, (CIN/COUT) ~ 2.4 (Figures 3-7).
Binding of polycyclic derivative to native E channel
Polycyclic derivatives of leading compound DABCO consists of a large number of atoms and can have large energy of interactions with the inside surface of E channel, PDB code 5X29. A set of molecular blockers considered here consists of larger number of atom, than DABCO. The first two molecules are diazabicyclooctan-benzene (DABCOB) and diazabicyclooctan-naphthalene (DABCON) in Figures 4 and 6 [1]. The second one are a two more complicated molecules with larger number of atoms and more voluminous molecule and 3D structure with equal size in three directions, namely, molecules diazabicyclooctan-3-benzene (DABCO3B) and diazabicyclooctan-3-toluene (DABCO3N) in Figures 5 and 7 [1]. It should be noticed, that construction of inhibitor of E channel using a rigid polycyclic skeleton is preferable compare to that using of a flexible frame, owing to a smaller loss of conformational entropy due to a restriction of conformational mobility of the drug molecule when it is bound inside the E channel. The results of docking of bicyclic and tricyclic derivatives of DABCO onto the E protein are shown in table and figures below [1]. It should be noted that residue GLU8 and ASN15 of different chains A, B, C, D, E are making a strong hydrogen bonds wit h drug molecules considered here (Table 2 and Figures 8-12).
| Drug moleculea | ePLb | eVDWc | eCould | eHbe | XHf∙∙∙Yg/a.a. Dist, Å | eSolvh | in(out)i |
|---|---|---|---|---|---|---|---|
| DABCO | -24.3 | -8.1 | -2.9 | -9.9 | GLU8A-OE2∙∙∙HN1 1.91 | -4.4 | in |
| GLU8E-OE1∙∙∙HN2 2.00 | |||||||
| -23.8 | -10.2 | -0.3 | -9.8 | VAL24C-O∙∙∙HN2 2.09 | -3.5 | out | |
| LEUE65-OXT∙∙∙HN1 1.94 | |||||||
| -23.5 | -11.8 | 4.9 | -9.7 | GLU8A.-OE1∙∙∙HN1 1.92 | -5.2 | in | |
| ASN15A-OD1∙∙∙HN2 2.06 | |||||||
| DABCOB | -33.5 | -13.8 | -7 | -9.9 | LEU9C-OXT∙∙∙HN2 1.97 | -2.7 | out |
| VAL47C-O∙∙∙HN1 1.98 | |||||||
| -34.4 | -13.9 | -6.9 | -9.9 | LEU65B-O∙∙∙HN1 2.09 | -3.7 | out | |
| VAL47C-O∙∙∙HN2 2.03 | |||||||
| -23.6 | -16.4 | -0.8 | -9.7 | ASN15C-OD1∙∙∙HN1 2.14 | -6.4 | in | |
| GLU8E-OE1∙∙∙HN2 2.08 | |||||||
| DABCO3B | -46.5 | -28.4 | -4.9 | -8.9 | ASN15C-OD1∙∙∙HN1 2.12 | -3.2 | in |
| GLU8E-OE1∙∙∙HN2 2.25 | |||||||
| -44.9 | -31 | -3.7 | -9.9 | LEU65A-OXT∙∙∙HN1 2.10 | -0.2 | out | |
| LEU21C-O∙∙∙HN2 2.18 | |||||||
| -44.4 | -28.6 | -5.3 | -9.6 | LEU65B-OXT∙∙∙HN1 2.01 | -0.9 | out | |
| VAL47C-O∙∙∙HN2 1.98 | |||||||
| DABCON | -38.8 | -16 | -3.2 | -9.7 | GLU8A-OE2∙∙∙HN1 2.06 | -9.9 | in |
| ASN15C-OD1∙∙∙HN2 2.14 | |||||||
| -35.9 | -17.3 | -4.4 | -9.9 | PHE20D-O∙∙∙HN1 2.07 | -4.3 | out | |
| LEU65C-OXT∙∙∙HN2 1.93 | |||||||
| -35.9 | -24.1 | 2.9 | -9.7 | SER50B-O∙∙∙HN1 2.12 | -4.9 | out | |
| TYNB-OH∙∙∙HN2 2.08 | |||||||
| DABCO3N | -58.1 | -34.1 | -6.8 | -9.1 | ASN15C-OD1∙∙∙HN1 2.19 | -8.1 | in |
| GLU8E-OE1∙∙∙HN2 2.15 | |||||||
| -52.3 | -38.1 | -3.4 | -5 | LEU65C-OXT∙∙∙HN1 2.01 | -5.8 | out | |
| -51.9 | -40.1 | -0.2 | -4.8 | GLU8A-OE1∙∙∙HN1 2.03 | -5.9 | out |
Note: aType of blocker molecule; bTotal energy of ligand binding by the E protein; cEnergy of van der Waals interactions; dEnergy of columbic interactions; eTotal H-bond energy; fH-bond donor, gH-bond acceptor; hInteraction with the solvent, kcal/mole; iDrug binding position, in or out of E channel.
Table 2: Binding positions ordered by binding energy of DABCO derivatives with E protein of SARS-CoV-2 virus, Glu8-Asn15–mut0.
Binding of drugs to mutant structures of E channel
Two the most probable mutating amino acid residues of E protein are mutations ASN15/ASP15 and GLU8/GLN8, with frequency of mutation equal to 42*10-4 and 35*10-4, respectively, as it is defined for a large set of protein mutants [37].Two single residue mutants of native E channel are considered, mut1 has mutation ASN15/ ASP15 in all fine chains (A, D, C, D, E) of protein E, and second one mut2 with mutation GLU8/GLN8 in all five chains. Mutants of E proteins have shown that the mutant structures are stable in a long runs MD simulation, Figure 2. The average energies over MD trajectories of the both mutant structures are higher compare to that of native E protein. The average energy of mut1 is higher by 30.1 and energy of mut2 is higher by 94.4 kcal/mole compare to the native structure. Taking into account that the rate of thermal fluctuations of potential energies over MD trajectories are 53.2 (for mut0), 50.1 (mut1), 47.3 (mut2), kcal/mole, it can be concluded that large enough amplitudes of thermal fluctuations of potential energies of mut0 and mut1 structures suggests, that mutants mut0 and mut1 can be convert one into another with high probability. Consider in detail the list of the main binding modes and its energies for five drug molecules, DABCO, DABCOB, DABCO3B, DABCON, DABCO3N in Figure 3-7 [1], with mutant, mut1, E channel are listed in the Table 3, the first three, the most strongest binding positions are the most strongest mode for each drug molecule which are shown in Figure 3-7. First of all, it should be noted that the strongest binding mode for all five drug molecules are in-channel binding positions (Table 3).
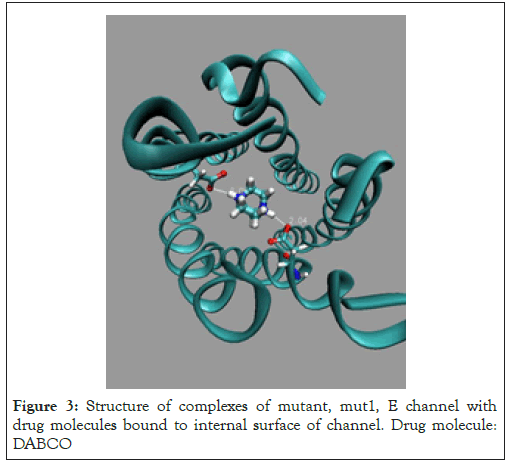
Figure 3: Structure of complexes of mutant, mut1, E channel with drug molecules bound to internal surface of channel. Drug molecule: DABCO

Figure 4: Structure of complexes of mutant, mut1, E channel with drug molecules bound to internal surface of channel. Drug molecule: DABCOB.

Figure 5: Structure of complexes of mutant, mut1, E channel with drug molecules bound to internal surface of channel. Drug molecule: DABCO3B.
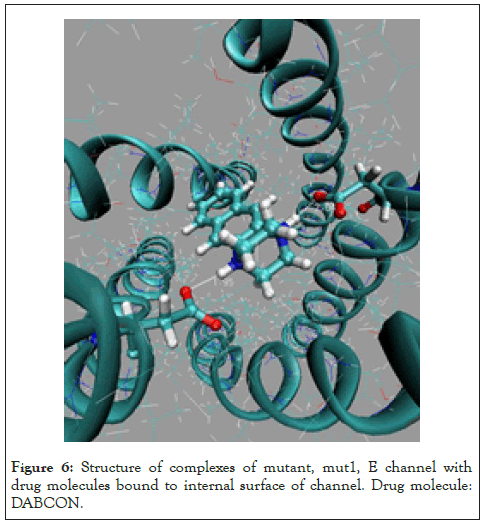
Figure 6: Structure of complexes of mutant, mut1, E channel with drug molecules bound to internal surface of channel. Drug molecule: DABCON.
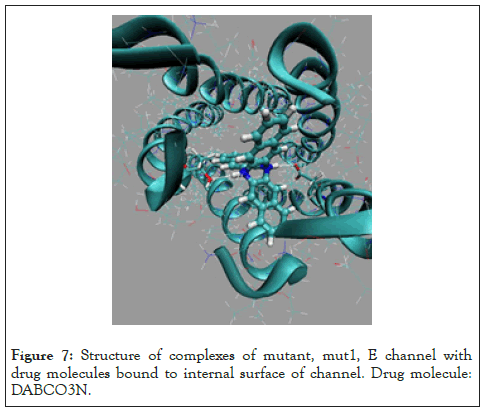
Figure 7: Structure of complexes of mutant, mut1, E channel with drug molecules bound to internal surface of channel. Drug molecule: DABCO3N.
| Drug moleculea | ePLb | eVDWc | eCould | eHbe | XHf∙∙∙Yg/a.a. | Dist, Å | eSolvh | in(out)i |
|---|---|---|---|---|---|---|---|---|
| DABCO | -36.5 | -10.2 | -15.1 | -9.8 | ASP15A-OD1∙∙∙HN1 | 2.04 | -1.4 | in |
| ASP15C-OD2∙∙HN2 | 2.06 | |||||||
| -35.3 | -10.5 | -12.7 | -10 | ASP15A-OD2∙∙∙HN1 | 1.88 | -1.1 | in | |
| ASP15C-OD1∙∙∙HN2 | 2 | |||||||
| -31.2 | -15.1 | -6 | -9.8 | ASP15A-OD1∙∙∙HN1 | 2.05 | 0.3 | in | |
| THR11B-OG1 HN2…………. | .2.06 | |||||||
| DABCOB | -43 | -15.8 | -16.8 | -9.9 | ASP15A-OD1∙∙∙HN1 | 1.9 | -0.8 | in |
| ASP15C-OD1…HN2 | 2.04 | |||||||
| -38.4 | -18.9 | -8 | -10 | ASP15A-OD1∙∙∙HN1 | 1.93 | -1.4 | in | |
| THR11D-OG1 HN2 | 2.04 | |||||||
| -36.6 | -18.5 | -10.2 | -8.6 | ASP15B-OG1…HN2 | 1.99 | 0.7 | in | |
| LEU12A-O…HN1 | 2.26 | |||||||
| DABCO3B | -56.6 | -32.3 | -11 | -9.7 | ASP15A-OD2∙∙∙HN1 | 2.16 | 2.8 | in |
| ASP15D-OD1…HN2 | 2.05 | |||||||
| -50.2 | -29.8 | -17.4 | -9.4 | ASP15A-OD1∙∙∙HN1 | 1.94 | 3 | in | |
| THR11D-OG1∙∙∙HN2 | 2.11 | |||||||
| -46.5 | -32.9 | -4.9 | -9.7 | ASP15A-OD1∙∙∙HN1 | 2.02 | 1.3 | in | |
| THR11B-O∙∙∙HN2 | 2.1 | |||||||
| DABCON | -45.2 | -19.7 | -16.1 | -9.9 | ASP15A-OD1∙∙∙HD1 | 1.94 | 0.6 | in |
| ASP15C-OD1∙∙∙HN2 | 2.12 | |||||||
| -42.8 | -19.3 | -11.9 | -10.1 | ASPA15-OD1∙∙∙HN1 | 1.95 | -1.1 | in | |
| ASP15C-OD1∙∙∙HN2 | 2.04 | |||||||
| -38.6 | -23.5 | -5.7 | -9.1 | ASP8-OD1∙∙∙HN1 | 1.95 | -0.3 | in | |
| LEU63-O∙∙∙HN2 | 2.12 | |||||||
| DABCO3N | -61.5 | -47 | -5.3 | -8.9 | ASP15A-OD2∙∙∙HN1 | 2.12 | 0.7 | in |
| THR11D-OG1∙∙∙HN2 | 2.23 | |||||||
| -52.4 | -45 | -9.8 | -4.8 | ASP15A-OG1∙∙∙ HN1 | 2.04 | 2.3 | in | |
| THR11B-O ∙∙∙HN2 | 2.4 | |||||||
| -57.8 | -54.1 | 3.4 | -9.1 | SER60D-OG∙∙∙HN1 | 2.21 | 2 | in | |
| ALA43A-O∙∙∙HN2 | 2.15 |
Note: aType of blocker molecule; bTotal energy of ligand binding by the E protein; cEnergy of van der Waals interactions; dEnergy of columbic interactions; eTotal H-bond energy; fH-bond donor, gH-bond acceptor; hInteraction with the solvent, kcal/mole; idrug binding position, in or out of E channel.
Table 3: Binding positions ordered by binding energy of DABCO derivatives with mutant Asn15/Asp15 of E protein of SARS-CoV-2 virus, Glu8- Asp15–mut1.
The respective binding energies are noticeably large ~5-11 kcal/ mole for the ASN15/ASP15 mutant, this is understandable because of positive charged DABCO derivatives have preferable electrostatic interactions with negatively charge amino acids of mutant, mut1, E channel. The preferable binding sites are characterized by formation of two strong hydrogen bonds between two nitrogen atoms of DABCO fragment of drug molecule with OD1/OD2 atoms of ASP15 of two different α-helixes. Table 3 shows that drug molecules DABCO3B and DABCO3N are the strongest binders to the inside surface of E channel. The second mutant of E channel considered here is mut2, GLU8/GLN8, in all five chains A, B, C, D, E. This mutation has replaced charged acid residues GLU8 to the neutral one GLN8. MD simulation of mutant GLU8/GLN8 in all five chains (A, B, C, D, E) of E protein, shows that the total potential energy of structure mut2 has the highest value among native and two mutant structures of E protein channel. The average energy of mut2 structure is large by ~60 kcal/mole compare to the average energy of mut1 and large by ~90 kcal/mole, relative to native sequence of E protein, Figure 2. The list of the main three, energy preferable, binding modes, and its energies for five drug molecules, DABCO, DABCOB, DABCO3B, DABCON, DABCO3N, with mutant, mut2, of E channel are listed in the table below. The most preferable binding modes are shown for each, of five, drug molecule in Figures 8-12 and Table 4.
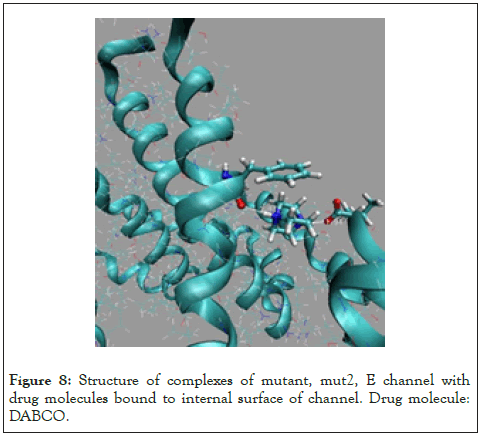
Figure 8: Structure of complexes of mutant, mut2, E channel with drug molecules bound to internal surface of channel. Drug molecule: DABCO.
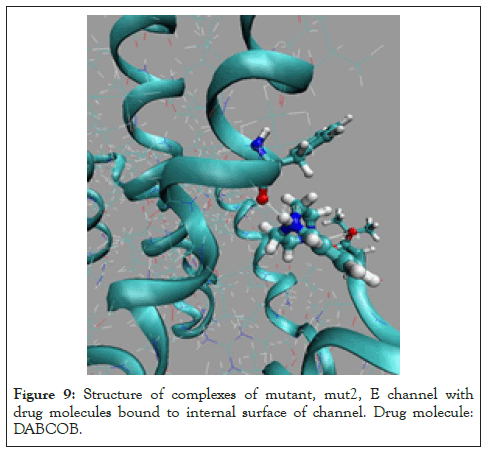
Figure 9: Structure of complexes of mutant, mut2, E channel with drug molecules bound to internal surface of channel. Drug molecule: DABCOB.
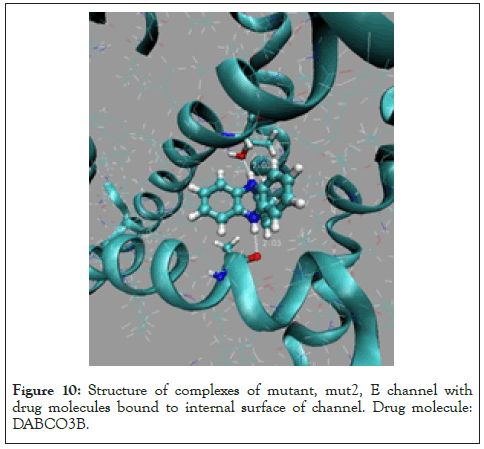
Figure 10: Structure of complexes of mutant, mut2, E channel with drug molecules bound to internal surface of channel. Drug molecule: DABCO3B.
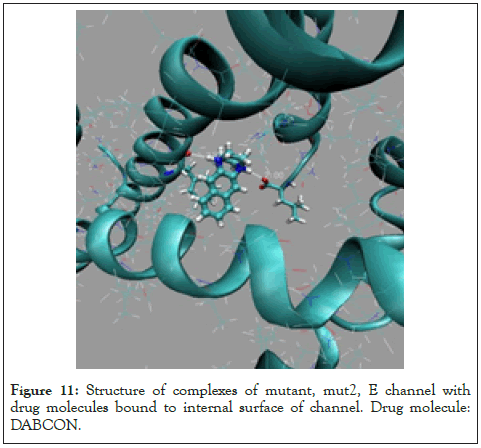
Figure 11: Structure of complexes of mutant, mut2, E channel with drug molecules bound to internal surface of channel. Drug molecule: DABCON.
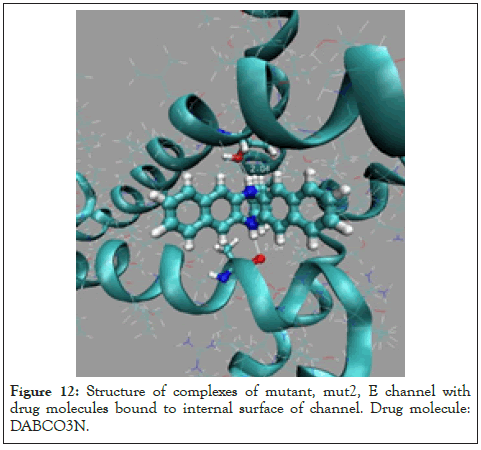
Figure 12: Structure of complexes of mutant, mut2, E channel with drug molecules bound to internal surface of channel. Drug molecule: DABCO3N.
| Drug moleculea | ePLb | eVDWc | eCould | eHbe | XHf∙∙∙Yg/a.a. | Dist, Å | eSolvh | in(out)i |
|---|---|---|---|---|---|---|---|---|
| DABCO | -25.6 | -17.7 | 8.1 | -9.8 | THR35C-OG1∙∙∙HN1 | 2.1 | -6.8 | out |
| ASN45B-OD1∙∙∙HN2 | 2.1 | |||||||
| -25.3 | -11.9 | 0.1 | -9.7 | LEU65C-OXT∙∙∙HN1 | 1.9 | -3.8 | out | |
| PHE20D-O∙∙∙HN2 | 2.1 | |||||||
| -18.5 | -13.6 | 8.3 | -9.5 | ASN15B-OD1∙∙HN1 | 2.1 | -3.7 | in | |
| ASN15C-OD1∙∙∙HN2 | 2.1 | |||||||
| DABCOB | -32.9 | -14.7 | -4.3 | -9.7 | PHE20D-O∙∙∙HN1 | 2.2 | -4.3 | out |
| LEU65C-O∙∙∙HN2 | 1.9 | |||||||
| -32.4 | -18.9 | 2.5 | -9.8 | ASN45B-OD1∙∙∙HN1 | 2 | -6.4 | out | |
| THR35C-O∙∙∙HN2 | 2.2 | |||||||
| -29.4 | -19.6 | 3 | -9.6 | ASN15C-OD1∙∙∙HN1 | 2 | -2.7 | in | |
| ASN15B-OD1∙∙∙HN2 | 2.1 | |||||||
| DABCO3B | -43.6 | -35.3 | 0.8 | -9.4 | ALA24E-O∙∙∙HN1 | 2 | 0.2 | out |
| THR30D-OE1∙∙∙HN2 | 2 | |||||||
| -40.7 | -33.5 | 1.3 | -8.2 | LEU37D-O∙∙∙HN1 | 2.3 | -0.6 | out | |
| ASN45C-OD1∙∙∙HN2 | 2 | |||||||
| -31.1 | -24.1 | 5.7 | -9.8 | GLN10A-O∙∙∙HN1 | 2.1 | -2.9 | in | |
| THR9B-OG1∙∙HN2 | 2.2 | |||||||
| DABCON | -37.4 | -21.1 | -1.8 | -9.8 | PHE20D-O∙∙∙HN1 | 2.1 | -4.7 | out |
| LEU65C-OXT∙∙∙HN2 | 2 | |||||||
| -33.8 | -18.8 | 0.2 | -9.8 | LEU65B-O∙∙∙HN1 | 1.9 | -5.4 | out | |
| THR55C-OG1∙∙∙HN2 | 2.1 | |||||||
| -33.9 | -25.7 | 2.4 | -9.7 | ASN15C-OG1∙∙∙HN1 | 2.1 | -0.9 | in | |
| ASN15C-OG1∙∙∙HN2 | 2.1 | |||||||
| DABCO3N | -54.8 | -50.9 | 2.4 | -5 | ASN64C-OD1∙∙∙HN1 | 2 | -1.4 | out |
| -54.6 | -45.3 | 1.5 | -9.3 | ALA32E-O∙∙∙HN1 | 2 | -1.5 | in | |
| THR30D-OG1H∙∙∙N2 | 2 | |||||||
| -53.7 | -40.4 | 1.9 | -9 | ASN45C-OD1∙∙∙HN1 | 2.1 | -6.2 | out | |
| THR35C-OG1∙∙∙HN1 | 2.3 |
Note: aType of blocker molecule; bTotal energy of ligand binding by the E protein; cEnergy of van der Waals interactions; dEnergy of columbic interactions; eTotal H-bond energy; fH-bond donor, gH-bond acceptor; hInteraction with the solvent, kcal/mole; iDrug binding position, in or out of E channel.
Table 4: Binding positions ordered by binding energy of DABCO derivatives with mutant GLU8/GLN 8 of E protein of SARS-CoV-2 virus, Gln8Asn15–mut2.
Table 4 shows that mut2 structure of E channel demonstrates that preferable binding mode of all five drug molecules are an outof- channel binding mode. It can be noted that the only blocker molecule, DABCO3N, demonstrate a good in-channel, binding with the binding energy practically equivalent to the best out-ofchannel mode. Analysis of data of the mut2, presented in the Table 4, and its comparison with data for negatively charged, native, charge of -1 e.u. and mut1, charge of -2 e.u., of the E protein structure, demonstrate that binding of molecular blockers are less strong with neutral mutant mut2. Analysis of data presented in Tables 2-4 suggests that positively charged molecules of five blockers DABCO, DABCOB, DABCO3B, DABCON, DABCO3N shows an importance of an effective electrostatic interaction with native and mut1 structures of E ion channel. Taking in mind that an electric neutrality of the entrance into the mut2 E channel is a negative factor, compare to that for the negatively charged entrance with GLU8 (A, B, C, D, E) in the native structure, therefore a diffusion rate of the positively charged ions H+/K+ will be decreased, compare to that for the native structure of the channel.
The paper describes results of construction of drug compounds, which can serve as an effective blocker of H+/K+ transport through cell membrane of virus SARS-CoV-2 . The wild native structure of E ion channel and mutant structures are considered. A long, more than 100 ns MD simulation of the native E protein and its two the most probable mutant structures, embedded into a lipid membrane were conducted at physiological conditions. It is found that the average structure and potential energies of native, mut0, PDB code 5X29, and mutants, mut1 ASN15/ASP15, and mut2 GLU8/GLN8 replacement for each of five chains of native 5X29 structure, in overall, have a similar structure, RMSD with native structure does not exceed 2.4 Å. The average potential energy difference between native, mut0 and mut1 structures are less than average amplitude of its thermal fluctuations at physiological conditions. Consequently, the probability of conversion mut0 ↔ mut1 is high. The potential energy of mut2, neutral, uncharged E channel structure is higher by ~90 kcal/mole, so that the probability of spontaneous conversion mut0→mut2 is low. The five drug molecules are suggested as molecular blockers of H+/K+ transport through E channel. It is shown that suggested compounds are an effective blocker of native mut0 and mut1mutant (ASN15/ ASP15) of E ion channel. Mutant GLU8/GLN8 of the E channel makes the channel entrance as neutral one. The role of electrostatic interactions of H+/K+ ions with the channel diminish and the probability of capture and diffusion of H+/K+ ions trough channel decreases. The most effective blocker compound which effectively bounds to the inside surface of native and mutants mut1, mut2 E channel and block H+/K+ ions transport through is the suggested compound diazabicyclooctan-3-toluene (DABCO3N).
This research was supported by Russian-Government–funded budget project for the Institute of Chemical Biology and Fundamental Medicine of the Siberian Branch of the Russian Academy of Sciences: #АААА-А17-117020210022-4.
[Crossref].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar].
[Crossref] [Google Scholar].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar].
[Crossref] [Google Scholar].
[Crossref] [Google Scholar].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar].
Citation: Vorobjev YN (2022) Molecular Blocker of Native and Mutant E Protein Ion Channel of Sars-Cov-2 Virus. Drug Des. 11:205.
Received: 25-Mar-2022, Manuscript No. DDO-22-16149; Editor assigned: 28-Mar-2022, Pre QC No. DDO-22-16149 (PQ); Reviewed: 12-Apr-2022, QC No. DDO-22-16149; Revised: 19-Apr-2022, Manuscript No. DDO-22-16149 (R); Published: 26-Apr-2022 , DOI: 10.35248/2169-0138.22.11.205
Copyright: © 2022 Vorobjev YN. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.