Advanced Techniques in Biology & Medicine
Open Access
ISSN: 2379-1764
ISSN: 2379-1764
Research - (2022)Volume 10, Issue 10
Mitochondrial RNAs are a class of small RNAs which can regulate gene expression through interference of translation process. A particular class of these miRNAs was found to be enriched in proximity of mitochondria and inside these organelles controlling the behavior of the mitochondria. The RNA in mitochondria is organized by special and membrane less compartments of RNA-protein complexes, called the RNA mitochondrial granules, which are abbreviated into MRGs.
Most methods used to isolate mitochondria depends on differential centrifugation, a two-step centrifugation carried out at low speed to remove intact cells, cell and tissue debris as well as nuclei from whole cell extracts followed by high speed centrifugation to concentrate mitochondria and separate mitochondria from other cellular organelles. Molecular quality control marker Hsp60 and organ-cellular quality control marker Atg5. Drug treatment with 6-OHDA induced decreased cell viability and ATP production of HtrA2 KO/CHOP genotype. Displaying a dmg concentration-dependent effect which reflects both the cell viability and the metabolic status of the cells. Drug treatment with 6-OHDA in conjunction with ADEP4 or ACP5 significantly improved cell survival, ATP production. Indicating that these drugs might have a protective effect while reoccurring the detrimental effects or 6-OHDA. The findings provide new evidence that depiction of HtrA2 might contribute to an increase in mitochondrial stress and transcriptional up regulation of the nuclear stress related to CHOP gene. Both are involved in Parkinson's disease.
RNAs; Mitochondria; Cellular organelles; Genotype
1960s chemical differences in the brains of PD patients identified. Levodopa was first administered and is still ‘gold standard medication: Arvid Carlsson, Oleh Hornykiewicz, George Cotzias, and Melvin Yahr won Nobel Prize in 2000 for their studies initiated with reserpine induced Parkinsonism in animal models.
Mitochondria are highly regulated membrane-bound organelles involved in fundamental cellular and metabolic functions within eukaryotic cells, such as energy supply (ATP production) through Oxidative Phosphorylation (OXPHOS), calcium and metabolites supply, inflammation, intracellular signaling, cell proliferation and apoptosis [1].
Cellular overexpression of HtrA2 results in variations of mitochondrial morphology and consecutive impairment and amplified susceptibility to stress –induced apoptosis. Depletion of HtrA2 can decrease mitochondrial level and mitochondrial membrane efficiency, which eventually results in ATP depletion and neuronal degeneration [2]. Genomic wide studies have demonstrated that HtrA2 depiction leads to transcriptional upregulation of important nuclear stress-response genes implicated mitochondrial PQC mechanisms, including the transcription factor CHOP (C/EBP homologous protein), which is differentially regulated upon depletion of this protease [3].
Mice without functional HtrA2 mi-protease activity showed induced upregulation of CHOP, reduced mitochondrial function and premature ageing in brain cells, due to an increase in mtDNA deletions [4].
DDIT3 (DNA damage induced transcript 3) also known as GADD 153 (Growth arrest and DNA damage 153) or CHOP (C/EBP homologous protein), encodes one of the six identified basic leucine zipper (bZIP) transcription factors of the dimer forming C/EBP family which is highly expressed in the cerebral cortex of the brain [5]. CHOP is considered a key precipitator transcriptional factor that plays a significant role in the regulation of mitochondrial homeostasis. CHOP expression is achieved by the mtUPR and the Integrated Stress Response (ISR), a general stress response program characterized by its involvement in the modulation of proteomic biosynthesis which is activated under different forms of stress [6]. Based on the hypothesis that depiction of HtrA2 might contribute to an increasing mitochondrial stress and transcriptional up-regulation of the nuclear stress-response CHOP gene, the present study aimed to analyze through a laboratory and bioinformatics-based research the contribution of HtrA2 depletion to the transcriptional up-regulation of CHOP in Parkinson's disease. To address the role of HtrA2 and CHOP in transmission of stress signaling and consequent activation of mitochondria quality control we have monitored one marker of molecular quality control, namely Hsp60 and one of mitophagy, Atg 5 (component of the phagosome elongation machinery) under pharmacologic activation of a mitochondrial stress signaling. Bioinformatics analysis was performed using online prediction tools to explore and analyze conserved protein domains and active regions.
MTT cell proliferation assay
Overview: MTT assay is the most commonly used viability assay and it was first described by Tim Mosmann in 1983.
Principle: This colorimetric assay uses reduction of a yellow tetrazolium salt (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, or MTT) to measure cellular metabolic activity. Viable cells contain NADP-H-dependent oxidoreductase enzymes which reduce the MTT reagent to formazan, an insoluble crystalline product with a deep purple color. Formazan crystals are then dissolved using a solubilizing solution as isopropyl alcohol and absorbance is measured at 500-600 nanometers using a plate-reader. The darker the solution, the greater the number of viable, metabolically active cells.
Reagents: MTT solution: MTT (5 mg/ml) in 0.9%NaCl.
Acidified isopropanol: HCl (0.04 N) in absolute isopropanol.
Calculations: The percentage of relative viability is calculated using the following equation:
[Absorbance of treated cells/Absorbance of control cells)]* 100
For extracting RNA from mitochondria, total cellular RNA is extracted from each cell line using TRizol (Sigma) reagent according to the manufacture's protocol for adherent cells. Briefly, after removing media, cells are lysed directly on the culture dish with 0.5 ml of TRizol reagent. For phase separation, 100 pl of chloroform is added to the samples which are vigorously agitated for 15 seconds and incubated for 15 minutes At Room Temperature (ART), followed by centrifugation at 13,400 xg for 15 minutes at 4°C. After phase separation, the upper colorless phase containing the RNA is transferred to fresh labeled micro-tubes and mixed with 200 pl of ice-cold isopropanol, incubated for 15 minutes ART, and centrifuged at 13,400 xg for 15 minutes at 4°C. The obtained RNA pellet is washed by the addition of 600 pl of 75% (v/v) ethanol, followed by centrifugation for 15 minutes at 13,400 xg at 4°C. The pellet is washed again with 600 pl of 75% (v/v) ethanol and stored for 24 hours in the fridge. Samples are centrifuged for 15 minutes at 13,400 xg at 4°C, air dried ART, and RNA pellets are re-suspended in 30 pl of nuclease free bO.
The purity and concentration of extracted RNA are quantified by a Nano Drop Spectrophotometer (Thermo Scientific).
6-OHDA treatment indicated decreased cell viability of HtrA2 KO/CHOP genotype as the different genotypes analyzed in this research were treated with 50 µM of the neurotoxin 6-OHDA and 0.5 µM of ADEP4 and ACP5. Then the cell viability was determined by MTT Assay. Cell viability has shown a concentration-dependent reduction of the viability level of the different genotypes. When compared to vehicle control (DMSO), cell viability was mainly reduced in the HtrA2 KO/CHOP genotype. It appeared that depletion of HtrA2 significantly increased the susceptibility to 6-OHDA treatment (p<0.0001).
These results display a drug concentration-dependent effect in the cell viability of the genotypes, indicating that depletion of cellular content might be due to high concentrations of drug treatment with 6-OHDA. Therefore, the concentration of the drug will be proportional to the number of living cells.
Drug treatment with 6-OHDA in conjunction with ADEP 4 or ACP5 have illustrated that these drugs might have a protective effect on CHOP KO. When compared to the genotypes treated only with 6-OHDA, cell viability was slightly less suppressed in HtrA2/CHOP KO genotype treated in conjunction with ADEP4 or ACP5.
The concentration-dependent effect of 6-OHDA drug treatment in the decrease of HtrA2/CHOP cell content could also be microscopically visualized When compared to the other drug treatments , HtrA2/CHOP KO genotype had a significantly lower concentration of living cells than the one treated only with 6-OHDA. Drug treatment with 6-OHDA in conjunction, with ADEP4 or ACPs significantly improved cell survival (Figure 1).
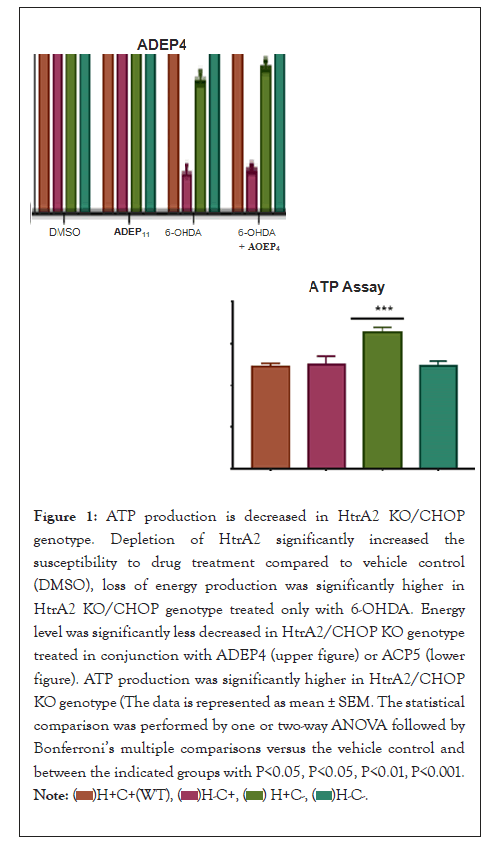
Figure 1: ATP production is decreased in HtrA2 KO/CHOP
genotype. Depletion of HtrA2 significantly increased the
susceptibility to drug treatment compared to vehicle control
(DMSO), loss of energy production was significantly higher in
HtrA2 KO/CHOP genotype treated only with 6-OHDA. Energy
level was significantly less decreased in HtrA2/CHOP KO genotype
treated in conjunction with ADEP4 (upper figure) or ACP5 (lower
figure). ATP production was significantly higher in HtrA2/CHOP
KO genotype (The data is represented as mean ± SEM. The statistical
comparison was performed by one or two-way ANOVA followed
by Bonferroni’s multiple comparisons versus the vehicle control
and between the indicated groups with P< 0.05, P<0.05, P<0.01,
P<0.001.
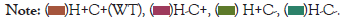
The HtrA2 gene comprises 4152 bases of DNA located in the forward strand of chromosome 2pl3.l and composed of eight exons (Figure 2). The gene encodes a 49 kDa protein of 458 amino acids (aa) residues. The full-length translated protein is located within the mitochondrial inter-membrane space (IMS) and contains a trimeric pyramidal structure which goes through a multifaceted allosteric activation pathway and defines HtrA2 proteolytic regulation, specificity and activity. The protease comprises the Nterminal mitochondrial localization sequence (1-40 aa), a transmembrane domain (10 5-125 aa) which dictates the topology of the protein, along with a serine protease domain (150-343 aa) with the catalytic triad residues (Ser306, His198 and Asp228), and finally one PDZ domain (364 -445 aa) located at the C-terminal end [7] (Figure 2).
Figure 2: Localization and schema tic 01•ganisation or the HTRA2 gene on chromosome 2p l3.1. The numbers indicate the length in kilo bases. Yellow boxes represent exons.
HtrA2 proteases are able to activate or coordinate heterogeneous signaling pathways implicated in PQC. Following apoptotic stresses, the N-terminus gets cleaved and the mature HtrA2 protein is released from the mitochondria Inter-Membrane Space (IMS) into the cytosol. Numerous structural studies have presented an overall analysis of the molecular architecture of human HtrA2, although the accurate pathway by which the protein is activated remains unclear [8].
HtrA2 family members belong to the corset of proteases which are found almost in all bacterial and eukaryotic genomes. In order to ascertain the evolution of the HtrA2 domain architecture, phylogenetic analysis using Phylogeny Software (www.phylogeny.fr) was performed. Phylogenetic tree analysis of the HtrA2 family members have indicated that HtrA2 from mouse (Mus musculus) was in the same subgroup of the other rodent (Rattus norvegicus). Based on high identity and similarity to human HtrA2 protein sequence, Mice Embryonic Fibroblasts (MEFs) are a good model of Parkinsonism to investigate the insight s of human PD.
In mitochondrial biogenesis, HtrA2 predominantly operates as a chaperone, where it prevents polypeptide aggregation and assists with correct protein folding. Although movement of the PDZ domain facilitated by s tress conditions such as temperature increase or by PDZ-binding peptides, were suggested to trigger HtrA2 activity, which initiates its proteolytic function and degrades the impaired proteins, similar to the allosteric mechanism suggested for the regulation of its bacterial homologue (HtrA2/DegP) [9].
Genomic wide studies have demonstrated that displacement or HtrA2 proteolytic activity results in accumulation of unfolded polypeptides within mitochondria, damage or the mitochondrial respiration and elevated ROS yield leading to impairment of mitochondrial bioenergetics [10]. Additionally, depletion of HtrA2 proteasome leads to transcriptional upregulalion or important nuclear stress-response gets implicated in mitochondrial PQC mechanisms, including the transcription factor CHOP (CEBP homologous protein), which is differentially regulated upon depletion of this protease.
Bioinformatics research shows that when compared, bZIP domains of the CHOP protease are highly conserved in human and in different types of eukaryotic organisms. According to NCBI BLAST comparison, the mouse CHOP and human CHOP (Homo sapiens) protein sequences (obtained from the Protein Data Bank) have 88% identity and 91 % similarity along their full length. A Cluslal Omega multiple sequence alignment performed with CHOP protein sequences from different species showed high sequence conservation in the bZIP domains of the analyzed species (Figures 3 and 4). The bZIP domains present in CHOP are structurally similar in their DNA-binding and dimerization motifs.

Figure 3: Comparison of human DDIT3/CHOP protein sequence with several other eukaryotic species. Clustal Omega multiple sequence analysis showed high sequence conservation in the bZIP domains of the analyzed species. Asterisks (*) indicate the residues that are identical, colons (:) indicate the conserved substitutions, and periods (.) indicate the semi-conserved substitutions.
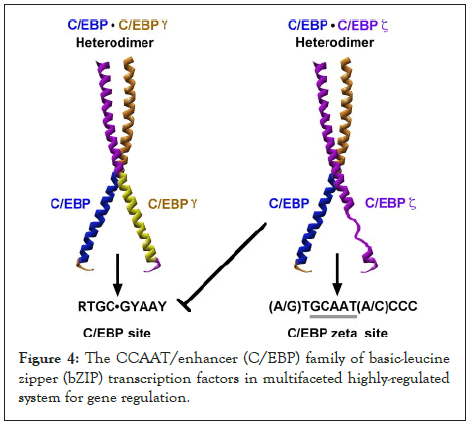
Figure 4: The CCAAT/enhancer (C/EBP) family of basic-leucine zipper (bZIP) transcription factors in multifaceted highly-regulated system for gene regulation.
CHOP hetero dimerizes with C/EBP and then attaches to the promoter region of the marker of molecular quality control, Hsp60, leading to the augmentation of its transcription levels. Atg5 is an important component of the phagosome elongation machinery. In vitro studies have indicated that cell animals lacking Atg5 suffered from depiction of neuronal cells. Therefore, removal of CHOP corroborates with the findings in the present research [11].
In response to removal of both HtrA2 and CHOP in cells, the results have indicated that this suppression might create a complex feedback loop mechanism that CHOP relays to in the mtUPR machinery, suggesting that CHOP is not functional to down regulate transcription. Upon the augmentation of cellular stress, CHOP/CEBP heterodimers have the capacity to attach to a different DNA sequence in the promoter regions of a sub-class of genes and trigger gene transcription. Accordingly, the CHOP/CEBP dimers bind and promote the activation of the promoters of the mtUPR responsive genes. Consequently, depending on the cellular state, CHOP can display both as an inhibitor of CEBP function and as a direct activator of other genes.
Cell viability analysis has shown a concentration-dependent reduction of the viability level of the different genotypes. When compared to vehicle control (DMSO), cell express ion was mainly down-regulated in the HtrA2 KO/CHOP genotype. It's appeared that depletion of HtrA2 significantly increased the susceptibility to dmg treatment, particularly to 6-OHDA (p<0.0001).
In the current research, the ATP assay displayed a concentration-dependent reduction of the energy production among the different genotypes. The results illustrated that depletion of HtrA2 significantly increased the susceptibility to dmg treatment. When compared to vehicle control (DMSO), loss of energy production was significantly higher in HtrA2 KO/CHOP genotype treated only with 6-OHDA (p<0.0001).
Total RNA can be also isolated from wide variety of tissues and cells using magnetic beads. Total RNA can be purified from 5 mg to 10 mg of tissue or from 5000 up to 1*106 cells. According to the results of these processes, new proofs were provided to explain that depiction of HtrA2 gene may participate in increasing mitochondrial stress and transcriptional up-regulation of the nuclear stress correlated with the activity of CHOP gene which is also involved in the role of early diagnosis and treatment of Parkinson's disease. RNA extraction and monitoring can help to identify the most part of mitochondria which is responsible to gene expression through interference with the gene translation process. A particular class of these mitochondrial RNAs was found to be enriched in proximity of mitochondria and within these organelles which play a vital role in controlling the mitochondrial behavior.
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
Citation: El-Sayed ABA (2022) Isolation of Mitochondrial RNA with 6-OHDA Treatment to Detect Cell Viability of HtrA2 KO/CHOP Genotype. Advanced Techniques in Biology & Medicine.10:380
Received: 19-Aug-2022, Manuscript No. ATBM-22-19817; Editor assigned: 22-Aug-2022, Pre QC No. ATBM-22-19817 (PQ); Reviewed: 06-Sep-2022, QC No. ATBM-22-19817; Revised: 13-Sep-2022, Manuscript No. ATBM-22-19817 (R); Published: 21-Sep-2022 , DOI: 10.35248/2167-1044.22.10.380
Copyright: © 2022 El-Sayed ABA. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.