Advanced Techniques in Biology & Medicine
Open Access
ISSN: 2379-1764
ISSN: 2379-1764
Mini Review - (2016) Volume 4, Issue 1
Due to the special affinity on tumor cells and anti-cancer activity for 5-iodo-4-thio-2'-deoxyuridine, the interactions between 5-iodo-4-thio-2'-deoxyuridine and human serum albumin (HSA) were investigated with the fluorescence spectroscopy. The results showed that 5-iodo-4-thio-2'-deoxyuridine on human serum albumin (HSA) had a dynamic fluorescence quenching. The main forces of both interactions have typical hydrophobic interaction from thermodynamic data to determination. The conformation of 5-iodo-4-thio-2'-deoxyuridine on human serum albumin was studied using synchronous fluorescence spectroscopy. The experimental result was in correspondence with molecular modeling theory. This work would be useful to understand the state of the transportation, distribution, and metabolism of the anticancer drug in human body, these results may be of significance in pharmacology and clinical medicines.
Keywords: 5-iodo-4-thio-2'-deoxyuridine, Human serum albumin (HSA), Nucleoside, Fluorescence spectroscopy, Molecular modeling
The application of modified nucleosides
Human serum albumin (HSA) is an important and the most abundant protein constituent of blood plasma and serves as a protein storage component. Recently, crystallographic studies of HSA have revealed that the protein, a 585 amino acid residue monomer, contains three homologous α-helical domains (I-III), and a single tryptophan (Trp214) [1]. HSA considerably contributes to colloid osmotic blood pressure and realizes the transportation and distribution of many molecules and metabolites such as fatty acids, amino acids, hormones, cations and anions, and many diverse drugs. It also makes possible to bind and carry many drugs through the bloodstream, which are poorly soluble in water [2]. Therefore, it is important to study the interaction of drug with the protein because protein-drug binding plays an important role in pharmacology and pharmaco dynamics. We can obtain a lot of information on the interaction between serum albumin and drug, which can help us better understand the absorption and distribution of the drug.
Nucleosides and their derivatives exhibit significant antitumor, antiviral and antibacterial activities [3]. It is recognized that nucleosides have the most potential function to restrain virus [4]. Extensive investigations into the interaction between the serum albumin and internal compound or pharmaceutical molecule have been made, but the interaction of protein with the nucleoside drug has seldom been reported [5]. In a series study methods concerning the interaction between drugs and protein, fluorescence techniques are great aids in the study of interactions between drugs and plasma proteins in general and serum albumin in particular because of their high sensitivity, rapidity, and ease of implementation [6], because such studies can provide information on the features affect the therapeutic effect of drugs. 5-iodo-4-thio-2'-deoxyuridine is one of important modified nucleosides, which oxygen atom was replaced by sulfur atom at 4-position in pyrimidine ring. In 2015, Zhang and Xu noticed that 5-iodo-4-thio-2'-deoxyuridine (4-SIdU) exhibited great anti-tumor activity for oral tumor which could kill cancer cells with UVA, so it can be a potential, nucleosides anti-tumor drug [7,8]. Therefore, it is of important worthiness and significance to investigate the interaction between 5-iodo-4-thio-2'-deoxyuridine and human serum albumin.
In this paper, we studied the interaction of 5-iodo-4-thio-2'- deoxyuridine with HSA at three temperatures under simulation of physiological conditions utilizing fluorescence in combination with molecular modeling. In the meantime, the binding mechanism ofs 5-iodo-4-thio-2'-deoxyuridine to HSA was discussed, and the binding parameters of the reaction were calculated according to fluorescence data.
The structure of 5-Iodo-4-thio-2'-deoxyuridine
The structure of 4-SIdU was showed in Figure 1. It is similar to the structure of 5-iodo-2'-deoxyuridine, with a thio atom instead of an oxygen atom at the C-4 position of the 2'-deoxyuridine. The pyrimidine containing thio carbonyl is redshift in maximum absorption peaks of UV and extremely sensitive to light. Therefore, a low dose near ultraviolet light can stimulate sulfur atom in thiocarbonyl for the electron transition which is destroying the structure of the compound 4-SIdU (Figure 1).
Materials and apparatus
UV-VIS spectrophotometer (JASCO, Japan); FP-6500 spectrofluorimeter (JASCO, Japan); pH-4 digital pH-meter (Shanghai Lei Ci Device Works, Shanghai, China); JULABO-F12 thermostat (Germany, ± 0.01°C); FA1004 Electronic balance (Shanghai Jing Ke Device Works, Shanghai, China)
Interaction with human serum albumin (HSA)
HSA (Sigma) was directly dissolved in double distilled water to prepare the stock solution (1.0 × 10-6 mol L-1 HSA), and the stock solution was kept in the dark at 0-4°C; 1.0 × 10-3 mol L-1 5-iodo-4-thio- 2'-deoxyuridin solution was obtained by dissolving it in double distilled water. 0.1 mol L-1 Tris-HCl buffer solution of pH 7.4, 0.1 mol L-1 NaCl working solution. Unless otherwise mentioned, all chemicals were of analytical reagent grade and were used without further purification. Double distilled water was used throughout the experiment.
UV absorption spectroscopy experiments of 5-iodo-4-thio-2'- deoxyuridine: Pipette 2ml concentration of 1 × 10-6 mol/L of human serum albumin solution to a 1cm quartz cuvette, Tris-HCl buffer solution as a reference, followed by adding an appropriate amount of micro-injector 5-iodo-4-thio-2'-deoxyuridine solution, allowed to stand for 3min, the UV absorption spectrum was measured at 220 ~ 450 nm (5-iodo-4-thio-2'-deoxyuridine solution was added a total volume of less than 50 μL).
Fluorescence spectroscopy experiments of 5-iodo-4-thio-2'- deoxyuridine: The quantitative analysis of the potential interaction between 5-iodo-4-thio-2'-deoxyuridine and HSA were performed by fluorescence titration (cumulative total volume of less than 50 μl). The reaction was carried out by mixing 2.5ml concentration of 1 × 10-6 mol/L of HSA stock solution with appropriate amount of 1.0 × 10-3 mol/L 5-iodo-4-thio-2'-deoxyuridine in the optical path of 10mm quartz cuvette, gently shaken, allowed to standing for 3 min. At 280 nm for the excitation wavelength of excitation and emission slit widths 5 nm/5 nm, the scanning speed was 500 nm/min, 0.05 mol/L Tris-HCl buffer solution as a blank correction. The fluorescence spectra of the range of HSA and HSA in the 5-iodo-4-thio-2'-deoxy-uridine were recorded by 290-450 nm on a fluorescence spectrophotometer under the action of the fluorescence quenching, 300 k, 310 k, and processing by the same method.
Synchronous fluorescence spectra: Synchronous Fluorescence Spectra were scanned on the same condition in section 4.2.2. The spectra were measured at two different em ex Δλ (Δλ = λem −λex) values, 15 nm and 60 nm.
Circular dichroism: Circular dichroism (CD) measurements were performed on a J-810 Spectropolarimeter (Jasco, Tokyo, Japan) at room temperature. CD measurements of HSA in the absence and presence of caffeine were recorded in the range of 260-200 nm. The instrument was controlled by Jasco’s Spectra Manager TM software. Quartz cells having path lengths of 0.1 cm were used at a scanning speed of 1000 nm/min. The data were expressed in terms of molar ellipticity, [θ]. An appropriate buffer solution run under the same conditions was taken as a blank and subtracted from the sample spectra.
Molecular modeling study
The initial structures of all the molecules were generated by molecular modeling software Sybyl 6.9.1. The potential of the 3D structures of HSA was assigned according to the Autodock 4.0 force field with Kollman-all- atom charges. The geometries of the antitumor drug (5-iodo-4-thio-2'-deoxyuridine) subsequently optimized using the Tripos force field with Gasteiger-Masili charges. The AutoDock 4.0 program was used to calculate the interaction modes between the drug and HSA. The Lamarckian Genetic Algorithm (LGA) applicated in AutoDock was applied to calculate the possible conformation of the drug that binds to the HSA. During the docking process, a maximum of 10 conformers was considered for the drug. The conformer with the lowest binding-free energy was used for further analysis. All calculations were performed on SGI FUEL workstations.
Interaction with human serum albumin (HSA)
UV-vis absorption spectra: In order to confirm the probable quenching mechanism, the UV-vis absorption spectra of 5-iodo- 4-thio-2'-deoxyuridine and HSA and the difference absorption spectrum {[HSA-5-iodo-4-thio-2'-deoxyuridine]-[5-iodo-4-thio-2'- deoxyuridine]} were also investigated (Figure 2). It can be seen from Figure 2 that the ultraviolet absorption peak between the 5-iodo-4- thio-2'-deoxyuridine and HSA was significantly different, while the ultraviolet absorption peak of HSA is almost no change by adding 5-iodo-4-thio-2'-deoxyuridine after from the experimental results, which confirms that the interaction between 5-iodo-4-thio-2'- deoxyuridine and HSA is mainly a dynamic quenching process [9].
Figure 2: The UV-vis absorption spectra of HSA in the absence and presence of 5-iodo-4-thio-2'- deoxyuridine. Curve B, the absorption spectrum of 5-iodo- 4-thio-2'-deoxyuridine only; curve D-M, the difference absorption spectrum between 5-iodo-4-thio-2'-deoxyuridin-HSA and 5-iodo-4-thio -2'-deoxyuridin, n (HSA): n (5-iodo-4-thio-2'-deoxyuridin)=1: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20.
Fluorescence spectra and quenching mechanism
Fluorescence quenching spectra study: The light from intrinsic fluorescence of the protein comes mainly from tryptophan, tyrosine and phenylalanine residues [10]. Due to quantum yield of phenylalanine is very low, where tyrosine is ionized or close to the amino and carboxyl group or tryptophan, and their fluorescence almost all quenched. The active sites in protein, HSA has only one tryptophan residue Trp214, the maximum emission wavelength at 332 nm, the intrinsic fluorescence of HSA is almost contributed by tryptophan residue, actually, tryptophan alone [11].
The fluorescence spectra of HSA at various concentration of 5-iodo-4-thio-2'-deoxyuridine are shown in Figure 3. Obviously, HSA has a strong fluorescence emission band at 332 nm while 5-iodo-4- thio-2'-deoxyuridine has no intrinsic fluorescence under the present experiment conditions when fixing the excitation wavelength at 282 nm. The fluorescence emission intensity of HSA decreases regularly with the increase of 5-iodo-4-thio-2'-deoxyuridine concentration. The strong quenching of HSA fluorescence clearly indicates that it have interacted with 5-iodo-4-thio-2'-deoxyuridine. The microenvironment around the Trp214 residue and the tertiary structure of HSA were changed after the addition of 5-iodo-4-thio-2'-deoxyuridine. There is an interaction between the HSA and 5-iodo-4-thio-2'-deoxyuridine.
Figure 3: The fluorescence emission spectra of HSA at 5-iodo-4-thio-2'- deoxyuridine various concentrations and an excitation wavelength of 282 nm in Tris-HCl butter solution (pH=7.4) at 27°C: Curve a, C (HSA) =6 μmol/L of the 5-iodo-4-thio-2'-deoxyuridine, curve b-h, the difference absorption spectrum between 5-iodo-4-thio-2'-deoxyuridin-HSA and 5-iodo-4-thio-2'-deoxyuridin, n (HSA) : n (4-SIdU) =1: 2, 4, 6, 8, 10, 12.
Quenching mechanism and binding constants: Fluorescence quenching refers to any process that decreases the fluorescence intensity of a sample. A variety of molecular interactions can result in quenching. It is important to recognize that the phenomenon of collisional quenching results in the expansion of the volume and distance within the solution which affects the fluorophore. The rootmean- square distance that a √Δx2. Fluorescence quenching process is usually divided into dynamic quenching and static quenching. The static quenching and dynamic quenching were differentiated by monitoring at different temperatures. The quenching rate constants decrease with increasing temperature for static quenching, but a reverse effect is observed for dynamic quenching [12]. In order to speculate the fluorescence quenching mechanism, the fluorescence quenching data at different temperatures (17, 27 and 37°C, Figure 4) were firstly analyzed using the classical Stern–Volmer equation (1) for dynamic quenching [13].
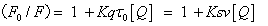 (1)
(1)
where F0 and F are the fluorescence intensities in the absence and presence of quencher respectively, Kq the biomolecular quenching constant, τ0 the life time of the fluorescence in absence of quencher, (Biological macromolecules of approximately τ0=10-8 S); in the absence of the presence of a quencher, the fluorescence lifetime of HSA τ0=1.77 × 10-8 S. [Q] the concentration of quencher (concentration of 5-iodo- 4-thio-2'-deoxyuridine), and KSV is the Stern–Volmer quenching constant, which is defined as the ratio of the bimolecular quenching rate constant and the decay rate constant of the single molecule (L/ mol). Therefore, it means that the mutual competition between these two decay pathway. As seen from Figure 4, F/F0 plotting [Q], the relative fluorescence intensity of the 332 nm at HSA according to equation (1) for dynamic processing, can be obtained through the slope of 5-iodo- 4-thio-2'-deoxyuridine of HSA quenching Ksv. obtained Kq (see Table 1). It was seen from Table 1, 5-iodo-4-thio-2'-deoxyuridine of HSA fluorescence quenching constant Kq is gradually increased as the temperature increased, and further description of such quenching process may be between molecules combined into a complex dynamic quenching.
| T(℃) | Stern-Volmer equation | Kq(L/mol) | R | Ea(KJ.mol-1) |
| 17 | (F0-F)/F=2.221×104[Q]-0.0127 | 2.221×1012 | 0.99686 | |
| 27 | (F0-F)/F=2.429×104[Q]-0.00461 | 2.429×1012 | 0.99979 | 7.36 |
| 37 | (F0-F)/F=2.705×104[Q]-0.00581 | 2.705×1012 | 0.99235 |
Table 1: The quenching constant (L/mol) and active energy Ea between 5-iodo-4- thio-2'-deoxyuridine and HAS by Stern-Volmer equation.
Both static and dynamic quenching required molecular contact between the fluorophore and quencher. One method to distinguish static and dynamic quenching is confirmed by careful examination of the absorption spectra of the fluorophore. To confirm dynamic quenching of interaction between 5-iodo-4-thio-2'-deoxyuridine and HSA, we also analyzed the fluorescence data at different temperatures with the well-known Lineweaver-Burk equations (2) for static quenching [14].
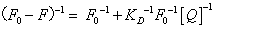 (2)
(2)
Where F0 and F are the fluorescence intensities in the absence and in the presence of quencher at concentration [Q]; and KD is the effective quenching constant for the accessible fluorophores.
For static quenching, the dependence of the fluorescence intensity upon quencher concentration is easily derived by consideration of the association constant for complex formation.
By Lineweaver-Burk double reciprocal ploting (F0-F)-1 on [Q]-1, it can be seen from Figure 5, curve linear relationship is good, the slope of the curve decreased as the temperature increased, respectively, by the reciprocal of the slope of the line can be determined at different temperatures of 5-iodo-4-thio-2'-deoxyuridine with human serum albumin in the dissociation constants and the correlation coefficient (see Table 2), as the temperature increases, the dissociation constant increases, the bonding between them is weakened. Thus, 5-iodo-4-thio- 2'-deoxyuridine can be stored and removed by protein in the human body.
| T(℃) | Lineweaver-Burk equation | KD(L/mol) | R |
| 17 | (F0-F)-1-F0-1= 61.1327 F0-1[Q]-1-0.0058 | 1.64×104 | 0.99727 |
| 27 | (F0-F)-1-F0-1=46.55492F0-1[Q]-1-0.00229 | 2.15×104 | 0.99915 |
| 37 | (F0-F)-1-F0-1=28.78507 F0-1[Q]-1+0.0053 | 3.47×104 | 0.97198 |
Table 2: The binding constants (L/mol) between 5-iodo-4-thio-2'-deoxyuridine and HSA by Lineweaver-Burk equation.
Analysis of binding equilibria: When small molecules bind independently to a set of equivalent sites on a macromolecule, the equilibrium between free and bound molecules is given by the equation (3) [15]:
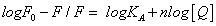 (3)
(3)
Where KA and n are the binding constant and number of binding sites, respectively. As can be seen from the Table 3, and KA and n are reduced with increased temperature, the phenomenon suggested that 5-iodo-4-thio-2'-deoxyuridine molecular bond formed unstable complexes with HSA, but as the temperature increases the complex formed. The complex begins to gradually decompose when the temperature increases.
| T(℃) | Scatchard equation | KA(L/mol) | n | R |
| 17 | 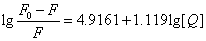 |
8.2413×104 | 1.12 | 0.99912 |
| 27 | 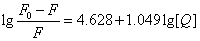 |
4.2461×104 | 1.05 | 0.9996 |
| 37 | 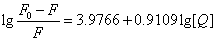 |
9.475×103 | 0.91 | 0.98218 |
Table 3: The binding constants (L/mol) and binding sites of 5-iodo-4-thio-2'- deoxyuridine and HSA by Scatchard equation.
Determination of the force acting between 5-iodo-4-thio-2’- deoxyuridine and HSA: The forces of interaction between drug and biomolecule may include hydrophobic forces, electrostatic interactions, van der Waals interactions, hydrogen bonding, etc. The slope of a plot of the bimolecular binding constant versus 1/T allows one to calculate the energy change for the quenching process [16]. Over a limited temperature range, the enthalpy change (ΔH) can be regarded as a constant, and its value and that of the entropy change (ΔS) can be determined from the van’t Hoff equation(4):
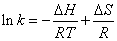 (4)
(4)
In this case, K is the binding constant Kb at the corresponding temperature. The temperatures used were 290K, 300K and 310K (Figure 6). The free energy change (ΔG) is estimated from the following relationship(5):
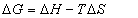 (5)
(5)
Where R is the gas constant, K is the binding constants at temperature. By the plotting the binding constants ln to 1/T (Figure 7).
Enthalpy change (ΔH) and entropy (ΔS) can be calculated by a straight line slope and intercept values, and thus according to the equation (6), free energy change (ΔG) can be calculated for the reaction.
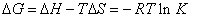 (6)
(6)
According to the above formula and the binding constants under different temperature, the thermodynamic parameters of the 4-SIdU and HSA system were calculated. As can be seen from Table 4, free energy change (ΔG) of 5-iodo-4-thio-2'-deoxy-uridine is negative, which indicates that 5-iodo-4-thio-2'-deoxy-uridine and HSA is a spontaneous process. The negative enthalpy (ΔH) and entropy (ΔS) values of the interaction of 5-iodo-4-thio-2'-deoxy-uridine and HSA, that is ΔH<0, ΔS<0, indicate that the binding is mainly enthalpy(ΔH)- driven and the entropy (ΔS) is unfavorable, This shows that the main role is hydrogen bonds and Van't der Waals forces between 5-iodo- 4-thio-2'-deoxy-uridine and HSA [16]. Hydrogen bonds are specific and directed and are probably best identified through negative of its enthalpy change (ΔH) value, we can exactly pointed out that the presence of hydrogen bonding between them.
| T(℃) | ΔG(kJ/mol) | ΔH(kJ/mol) | ΔS(J mol-1K-1) |
| 17 | -27.674 | ||
| 27 | -25.838 | -80.912 | -183.58 |
| 37 | -24.002 |
Table 4: Thermodynamic parameter of 5-iodo-4-thio-2'-deoxyuridine and HAS interaction.
Energy transfer between 5-iodo-4-thio-2'-deoxyuridine and HSA: Fluorescence resonance energy transfer (FRET) is a spectroscopic method that can monitor the proximity and relative angular orientation of fluorophores. The donor and acceptor fluorophores can be entirely separated or attached to the same macromolecule. The energy transfer can be replaced through direct electrostatic interaction between the initially excited molecule and its neighbors [17,18].
Förster dipole-dipole non-radiative energy transfer theory [19] that (1) the energy between the donor and acceptor molecules non-radiative energy transfer must meet: donor fluorescence emission spectrum and acceptor absorption spectra between appropriate overlap and the maximum spacing of no more than two molecules 7 nm; (2) the role of donor-acceptor distancer can be accurately calculated by the following formula (7) [20].
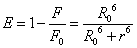 (7)
(7)
Where E denotes the efficiency of energy transfer between the donor and acceptor, and r is the distance between the donor and acceptor. R0, the critical distance at which the transfer efficiency equals 50%, is given by the eq. (8):
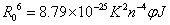 (8)
(8)
In eq. (9), K2 is the space factor of orientation, n is the refractive index of the medium, φ is the fluorescence quantum yield of the donor. J expresses the degree of spectral overlap between the donor emission and the acceptor absorption spectrum (Figure 8), which can be calculated by the eq. (9):
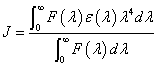 (9)
(9)
where, F(λ) is the corrected fluorescence intensity of the donor in the wavelength range λ to λ+Δλ; and ε (λ) is the extinction coefficient of the acceptor at λ. J could be evaluated by integrating the spectra in Figure 8. According to the reported for HSA in the literature K2=2/3, n=1.336, and φ =0.118 [21]. The value of the overlap intergral calculated from Figure 8 is J=3.66 × 10-14 cm3 L mol-1, and thus according to eqs. (8)-(10), we can calculate values of R0=2.76 nm and r=3.32 nm. When a fluorophore gives the average distance between the body and the fluorophore acceptor in the 2-8 nm range and meet 0.5 R0
Investigation of conformational changes on binding of 5-iodo-4- thio- 2'-deoxyuridine to HSA
Synchronous fluorescence spectroscopy studies: Synchronous fluorescence spectroscopy can give information about the molecular environment in the vicinity of the chromosphere molecules at low concentrations under physiological condition. It involves the simultaneous scanning of the excitation and the fluorescence monochromators of a fluorimeter, while maintaining a fixed wavelength difference (Δλ) between them [23]. Therefore, the conformation changes of HSA were evaluated by measuring the synchronous fluorescence intensity of protein amino acid and residues, both before and after the additions of complex. Amino acid residues of the maximum emission peak positions associated with the polarity of the microenvironment of the changes in protein conformation, it can be judged from the maximum emission peak bit changes: the maximum emission wavelength of blueshift that increased hydrophobicity of the amino acid residues in their environment, red-shift indicates that polarity increased [24]. When the Δλ=λem-λex between excitation and emission wavelength is fixed at Δλ=15 nm or Δλ=60 nm, the synchronous fluorescence gives characteristic information about the tyrosine or tryptophan residues, respectively [25,26]. It can be seen from Figure 9 that when Δλ was set at 15 nm (Figure 9 (left)), the maximum emission wavelength undergoes a more slight red-shift of 0.4 nm (from 300.4 nm to 300 nm) for the investigated concentration range; when Δλ=60 nm (Figure 9 (right)), the maximum emission wavelength shows a more red-shift of 0.2 nm (from 340.6 nm to 340.8 nm). The change of these data is so very a little that explains hydrophobicity or polarity of the amino acid residues in their environment.
It was found the degree of fluorescence quenching of the Tyr residues was greater than that of Trp residues by Synchronous fluorescence spectroscopy of residues in Tyr and Trp. This suggested that the 4-SIdU is more close to Tyr residues of HSA. The reason of 4-SIdU quenching Trp residues of HSA may be due to the 4-SIdU molecular motion around Trp214 residue of the HSA protein active sites suitable range, it is possible to quench off the fluorescent of Trp residues by way of dynamic quenching and energy transfer. In addition, it was also found that the maximum emission peak at position basically unchanged by the synchronous fluorescence spectra of the two amino acid residues, this suggests that the maximum emission peak description the 4-SIdU their microenvironment had little effect and did not lead to changes in the conformation of the protein molecule.
The CD spectra of interaction between HSA and 5-iodo-4-thio- 2'-deoxyuridine: The secondary structure of the protein molecules caused by some of the reactions was sensitively detected by the circular dichroism spectra. The circular dichroism spectra of interaction between HSA and 5-iodo-4-thio-2'-deoxyuridine are shown in Figure 9. There are two negative bands at 208 nm and 222 nm in the circular dichroism spectra, which is a typical α-helix structure of the CD spectra signal. A reasonable explanation is that the negative peaks between 208 nm and 222 nm both arise from n→π* transfer in the peptide bonds of the α-helix [27]. It can be seen from Figure 10 when the concentration of 5-iodo-4-thio-2'-deoxyuridine from 2.0 × 10-5 mol/L gradually increased to 1.4 × 10-4 mol/L, in other words, the binding of 5-iodo-4- thio- 2'-deoxyuridine to HSA decreases the intensities of both of these bands, clearly indicating a decrease in the α-helix content of the protein. The secondary structure of the protein molecules HSA to 5-iodo-4- thio-2'-deoxyuridine basically no change and the circular dichroism spectra also has not changed basically. This indicates that 5-iodo-4- thio-2'-deoxyuridine binds with the amino acid residues of the main polypeptide chain of the protein and destroys their hydrogen bonding networks [28]. The results showed that 5-iodo-4-thio-2'-deoxyuridine role in the process of tumor cells by blood affect the structure of HSA is a small, there are extremely weak changes in the secondary structure of HSA. These indicate that the kinds of anti-tumor drugs in the human body are almost no change in the secondary structure of HSA. Therefore, HSA is a kind of the carrier protein which can safely carry anti-tumor drugs (5-iodo-4-thio-2'-deoxyuridine) to reach the affected area of tumor cells [29].
Molecular modeling study of the interaction between HSA and 5-iodo-4-thio-2’-deoxyuridine
Some researcher have shown that HSA is able to bind many ligands in several binding sites [1,30-32]. From the 3D structure of crystalline albumin, there are three homologous (denoted?, ?and ?) included in the sites of HSA, such as,?comprises residues (1-195), ?(196- 383); and ? residues (384-585), each with two subdomains, A and B possessing common structural patterns. It is suggested that the principal regions of ligand binding to HSA are located in hydrophobic cavities in subdomains ?A and ?A, which are accordance with site?and ?, respectively, and one tryptophan residues (Trp-214) of HSA is in subdomain ?A [2]. There is a large hydrophobic cavity reflect in subdomain?A that many drugs can also bind. The crystal structure of HSA was taken from the Brookhaven Protein Data Bank (entry code 1H9Z) [32]. The potential of the 3D structure of HSA was assigned according to the Amber 4.0 force field with Kollman-all-atom charges. The initial structures of all the molecules were generated by molecular modeling software Sybyl 6.9.1 [33]. The geometries of these compounds were subsequently optimized using the Tripos force with Gasteiger-Marsili charges. The Autodock 4.0 program was used to calculate the interaction between the ligands and HSA [34,35]. The Lamarckian Genetic Algorithm (LGA) applicated in AutoDock was applied to calculate the possible conformations of the ligands that bind to the protein. During the docking process, a maximum of 10 conformers was considered for this compound. The conformer with (rootmeans-square) was used for method, a computational model of the target receptor has been built; Partial binding parameters of 5-iodo- 4-thio-2’-deoxyuridine-HSA complex system were calculated through SGI FUEL workstations.
The best energy ranked result is shown in Figure 11. In addition, Figure 12 shows the macroscopic viewer of the interaction between drug and HSA. It was evident that 5-iodo-4-thio-2’-deoxyuridine molecule was located in the subdomain?A hydrophobic cavity, and 5-iodo-4-thio-2’-deoxyuridine was adjacent to hydrophobic residues, such as Trp-214, Lys-199, Phe-211, Ser-202, Ala-210, Phe-206, Leu-347, Leu-481 of subdomain?A of HSA. It is suggested that the existence of hydrophobic interaction between them. Further, this finding provides a good structural basis to interpret the efficient fluorescence quenching of HSA emission in the process of 5-iodo-4-thio-2’-deoxyuridine. From the Figure 11, there is a weak hydrogen bond between 5-iodo- 4-thio-2’-deoxyuridine and the residues (Phe-206), so, the hydrogen bond is not displayed. The results obtained from modeling conformed that the interaction between 5-iodo-4-thio-2’-deoxyuridine and HSA was mainly dominated by hydrophobic force.
Based on a special affinity on tumor cells and anti-tumor activity for nucleoside compounds, 5-iodo-4-thio-2’-deoxyuridine was studied. An approach for studying the binding of 5-iodo-4-thio-2’-deoxyuridine to HSA was provided by employing different optical techniques such as fluorescence spectroscopy, UV-vis absorption spectroscopy and CD spectroscopy in this paper. The results of spectral peak intensity decreased in synchronous fluorescence and fluorescence calculation showed that 5-iodo-4-thio-2'-deoxyuridine role in distance tryptophan closer HSA active sites, forming unstable complexes with HSA through the static fluorescence quenching procedure, while the results of ultraviolet and circular dichroism indicate that protein structure was affected by the subtle role of 5-iodo-4-thio-2'-deoxyuridine which was confirmed by the results of molecular modeling. These results could provide a useful biochemistry parameter for the development of the new anticancer drugs and research of pharmacology mechanisms, and it would be of significance in pharmacology and clinical medicines.
The work is financially supported by the Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry, P. R. China. The project received funding as part of the Dalian government's science and technology plan projects [2014E12SF074], of Liaoning Province Education Administration [L2013472].