Organic Chemistry: Current Research
Open Access
ISSN: 2161-0401
ISSN: 2161-0401
Editorial - (2012) Volume 1, Issue 3
Site-selective protein labeling is an indispensible approach in the currently intense chemical biology research area. Studies involving site-selective protein labeling span from the protein dynamic analysis in vitro to the proteinprotein interaction investigation in living cells. In the past decade, multiple methods have been introduced to achieve site-selective protein labeling. These include genetic fusion of green fluorescent protein and its derivatives, selective chemical labeling of proteins with fusion tags, and site-specific modification of noncanonical amino acids that are genetically encoded. Using evolved orthogonal aminoacyl-tRNA synthetase-nonsense suppressor tRNA pairs, noncanonical amino acids with bioorthogonal functional groups such as azide, alkyne, tetrazine, alkene, keto, phenylhalide, etc. have been genetically incorporated into proteins in E. coli, yeast, and mammalian cells. Genetic encoding of these noncanonical amino acids enables multiple ways for site-selective protein labeling both in vitro and in vivo, allowing diverse strategies to interrogate protein functions. This review intends to provide a brief introduction to the genetic noncanonical amino acid incorporation technique and recent progresses in applying this technique to achieve site-selective protein labeling.
Keywords: Noncanonical amino acids; Nonsense suppression; Click chemistry; Bioorthogonal functional groups; Genetic code expansion
Protein labeling with fluorescent molecules that allows sensing and visualization of protein dynamics, localization, protein-ligand interactions, and protein-protein interactions, is an invaluable tool to understand protein functions in living cells. One of the most prominent methods of fluorescent protein labeling is to genetically encode green fluorescent protein (GFP) or one of its variants as a fusion to the protein of interest. This powerful technique has intrinsic important advantages such as high labeling specificity and simplicity [1-5]. In 2008, Nobel Prize in chemistry was awarded to three scientists Osamu Shimomura, Martin Chalfie and Roger Y. Tsien for their discovery and development of GFP, highlighting the great contribution of the GFP technique in advancing chemical and biological research. Although GFP variants have proved to be extremely useful for both in vitro and in vivo studies of protein functions, their utility is still limited because the molecular sizes of GFP variants (~27kDa) are large enough to potentially interfere with the structure and function of proteins to which they are fused and their spectral and structural characterization are interdependent [6-8]. To increase the diversity of protein labels, approaches comparable and complementary to the GFP technique have been developed that confer selectively fluorescent labeling of proteins with smaller chemical moieties.
Tag-based chemical labeling approaches have flourished recently. They require genetically fusing target proteins to peptide tags that specifically bind to or react with small molecule probes consisting of fluorophores. A great advantage of tag-based chemical labeling approaches is the flexibility in choosing fluorophores. A major advance in the tag-based protein labeling was achieved when biarsenical fluorescent dyes were used to label fusion proteins containing a tetracysteine (TC) motif [9-12]. Binding of a biarsenical dye, notably green-fluorescent FlAsH or red fluorescent ReAsH, to the TC tag forms a stable fluorophore-protein complex. The small size of the tag is a proven advantage in several direct comparisons with GFP variants [8,13,14]. Following the introduction of the TC tag, other tags for chemical labeling of proteins have been developed. They fall into two categories: peptide tags rationally designed or evolved for binding to chemical probes and peptide tags from natural biosynthetic pathways that serve as specific sites for covalent attachment of chemical groups by enzymes. The first category includes small fusion tags [15-19], DHFRtag that non-covalently binds fluorescent trimethoprim derivatives [20,21], SNAP-tag that covalently reacts with fluorescent O6-benzylguanine substrates [22-26], and halo-tag that cleaves the carbon halogen bond of fluorescent ligands to become covalently labeled with the fluorophores [27-29]. Labeling of peptide tags in the second category all involve enzymes. In the ACP-tag system, a phosphopantetheine transferase enzyme is used to transfer a 4’-phosphopantetheine-linked probe from coenzyme A to a serine residue of acyl carrier protein (ACP) that is fused to other proteins [30-35]. In another system, biotin ligase has been used for covalent labeling of a 15-aa peptide tag with ketone-modified biotin molecules that can react with hydrazide or hydroxylamine fluorescent dyes [36-38]. The use of formylglycinegenerating enzyme to generate formylglycine in a 13-aa peptide tag for labeling with hydrazide or hydroxylamine fluorescent dyes [39-41] and the exploitation of lipoic acid ligase to transfer an azide-containing lipoic acid probe to a 22-aa peptide tag for labeling with alkyne dyes [42,43] have also been successfully demonstrated. Although many tag-based chemical labeling techniques have advantages like flexibility in choosing fluorophores and less disturbance of structures and functions of target proteins, they have inherent limitations as well. First, a tag is generally fused to the N- or C-terminus of the protein of interest. Installation of a fusion tag at internal part of the protein without disrupting protein structures and functions is difficult. Second, although most fusion tags are considerably smaller than GFP variants (the TC tag has only six residues), they are still not single residues. In comparison to single-residue modification, these fusion tags more likely adversely affect structures and functions of proteins they are fused to.
There are a few methods available for labeling proteins at single residue levels. One broadly used approach exploits the reactivity of cysteine residues within proteins and labels them with appropriate thiol-reactive dyes [44]. Native chemical ligation and its extension, expressed protein ligation, have also been used to introduce various probes at single-residue levels [45-47]. These two approaches have, however, been generally limited to in vitro modification of purified proteins. Biological studies of such labeled proteins in cells require their reintroduction by invasive techniques such as microinjection or electroporation. Moreover, modification based on cysteine residues requires mutating all other non-targeted cysteine residues that could be critical to protein functions. For the native chemical ligation method, appropriate sites for ligation must be chosen carefully and modification of internal sites in large proteins is cumbersome. Therefore, to resolve issues associated with these aforementioned techniques, a singleresidue labeling approach that is easy to perform in vitro and allow non-invasive labeling of proteins in vivo is necessary.
Genetically incorporating NAAs into proteins is an alternative powerful approach that allows site-selective labeling of proteins at single-residue levels. A general method for the genetic NAA incorporation approach in live cells was developed by Furter, Schultz, and their coworkers [48-55]. This method relies on the read-through of an in-frame stop codon in mRNA by a nonsense suppressor tRNA that is specifically acylated with a NAA by an evolved aminoacyl-tRNA synthetase. There is a naturally existing genetic NAA incorporation system. In some methanogenic archaea and a Gram-positive bacterium Desulfitobacterium hafniense, Pyl is co-translationally inserted into proteins by an in-frame amber codon [56-61]. Suppression of this amber codon is mediated by the Pyl amber suppressor tRNA (  ), which has a CUA anticodon and is acylated with Pyl by pyrrolysyl-tRNA synthetase (PylRS). The PylRS-
), which has a CUA anticodon and is acylated with Pyl by pyrrolysyl-tRNA synthetase (PylRS). The PylRS-  pair in these organisms is orthogonal to other synthetase-tRNA pairs in cells, ensuring the fidelity of the Pyl incorporation. Similarly to the Pyl incorporation machinery, an orthogonal synthetase-nonsense suppressor tRNA pair can be developed in which the synthetase is evolved to specifically charge its cognate suppressor tRNA with a NAA. When expressed in cells, this orthogonal synthetase-suppressor tRNA pair enables the NAA to be site-specifically incorporated into a protein at the amber codon with high fidelity and efficiency. Using this approach, a variety of NAAs have been incorporated into proteins in bacteria, yeast and mammalian cells and used to study a large number of biological problems [48,62-66]. Three of these NAAs are fluorescent themselves [65,66]. Many others have chemically reactive groups such as phenylhalide, ketone, azide and alkyne, alkene, tetrazine, tetrazole, etc. These groups can be directly used to introduce fluorescent labels into proteins both in vitro and in vivo [62,63]. The genetic NAA incorporation approach for fluorescent protein labeling has essential advantages. First, it relies simply on the recombinant DNA technique so that it can be easily generalized. Large amount of modified proteins can be generated easily. Second, the labeling is site-directed and sitespecific regardless of the incorporation site or protein size. Third, the localizable labeling in live cells can be achieved using chemically reactive NAAs and fluorescent dyes that have varied permeability to different organelles.
pair in these organisms is orthogonal to other synthetase-tRNA pairs in cells, ensuring the fidelity of the Pyl incorporation. Similarly to the Pyl incorporation machinery, an orthogonal synthetase-nonsense suppressor tRNA pair can be developed in which the synthetase is evolved to specifically charge its cognate suppressor tRNA with a NAA. When expressed in cells, this orthogonal synthetase-suppressor tRNA pair enables the NAA to be site-specifically incorporated into a protein at the amber codon with high fidelity and efficiency. Using this approach, a variety of NAAs have been incorporated into proteins in bacteria, yeast and mammalian cells and used to study a large number of biological problems [48,62-66]. Three of these NAAs are fluorescent themselves [65,66]. Many others have chemically reactive groups such as phenylhalide, ketone, azide and alkyne, alkene, tetrazine, tetrazole, etc. These groups can be directly used to introduce fluorescent labels into proteins both in vitro and in vivo [62,63]. The genetic NAA incorporation approach for fluorescent protein labeling has essential advantages. First, it relies simply on the recombinant DNA technique so that it can be easily generalized. Large amount of modified proteins can be generated easily. Second, the labeling is site-directed and sitespecific regardless of the incorporation site or protein size. Third, the localizable labeling in live cells can be achieved using chemically reactive NAAs and fluorescent dyes that have varied permeability to different organelles.
One great advantage of the GFP labeling technique is its efficiency and simplicity. When expressed in cells, a self-catalyzed process generates the GFP fluorophore and ensues quantitative labeling of a target protein that is fused to GFP. On the contrary, most chemical labeling strategies require further treatment after protein expression and therefore are more complicated [67-71]. An ideal chemical labeling approach that can achieve comparable simplicity and efficiency of the GFP labeling technique is to directly incorporate fluorescent NAAs into proteins. As of today, three fluorescent NAAs shown in Figure 1 have been genetically incorporated into proteins. Using an evolved tyrosyltRNA synthetase (MjTyrRS)-amber suppressing  pair that was derived from Methanocaldococcus jannaschii, 1 was genetically installed into proteins in E. coli at amber mutation sites. 1 contains a fluorescent 7-hydroxycoumain moiety that shows a high fluorescent quantum yield, a relatively large Stoke’s shift, and sensitivity to pH and solvent polarity. These unique features of 1 have been applied to undergo a variety of protein function studies such as protein folding/unfolding, protein-protein interaction, and protein subcellular localization. Using myoglobin incorporated with 1, Schultz and coworkers showed the sensitivity of fluorescence of 1 to the polarity of its environment could be used to track the unfolding process of myoglobin incorporated with 1 in urea [66]. This same physical property of 1 has also been used to visualize antibody-antigen interactions and the phosphorylation state of STAT3 [72,73]. The strong fluorescence of 1 has also been used to investigate subcellular localization of GroEL and FtsZ in E. coli. FtsZ is a bacterial tubulin homologue. FtsZ incorporated with 1 is seen at the cleavage furrow during cell division to form the Z-ring, providing the first example of a fully functional protein to be visualized in living cells using a genetically incorporated NAA [74,75]. Another fluorescent NAA that has been incorporated into proteins is 2 in Figure 1 that contains a fluorescent dansyl functional group. The incorporation of 2 into proteins in yeast and mammalian cells was achieved using an evolved leucyl-tRNA synthetase (EcLeuRS)-amber suppressing
pair that was derived from Methanocaldococcus jannaschii, 1 was genetically installed into proteins in E. coli at amber mutation sites. 1 contains a fluorescent 7-hydroxycoumain moiety that shows a high fluorescent quantum yield, a relatively large Stoke’s shift, and sensitivity to pH and solvent polarity. These unique features of 1 have been applied to undergo a variety of protein function studies such as protein folding/unfolding, protein-protein interaction, and protein subcellular localization. Using myoglobin incorporated with 1, Schultz and coworkers showed the sensitivity of fluorescence of 1 to the polarity of its environment could be used to track the unfolding process of myoglobin incorporated with 1 in urea [66]. This same physical property of 1 has also been used to visualize antibody-antigen interactions and the phosphorylation state of STAT3 [72,73]. The strong fluorescence of 1 has also been used to investigate subcellular localization of GroEL and FtsZ in E. coli. FtsZ is a bacterial tubulin homologue. FtsZ incorporated with 1 is seen at the cleavage furrow during cell division to form the Z-ring, providing the first example of a fully functional protein to be visualized in living cells using a genetically incorporated NAA [74,75]. Another fluorescent NAA that has been incorporated into proteins is 2 in Figure 1 that contains a fluorescent dansyl functional group. The incorporation of 2 into proteins in yeast and mammalian cells was achieved using an evolved leucyl-tRNA synthetase (EcLeuRS)-amber suppressing  pair that was derived from E. coli. Like 1, fluorescence of 2 is also sensitive to solvent polarity. Genetic incorporation of 2 into proteins can be applied to study protein folding/unfolding processes [65]. By genetically incorporating 2 into a voltage-dependent membrane lipid phosphatase, Wang and coworkers showed that 2 optically reports the conformational change of the voltage-sensitive domain in response to membrane depolarization [76]. Another fluorescent NAA that has been genetically incorporated into proteins is 3. Like 1 and 2, 3 is also an environment-sensitive fluorescent NAA. Using an evolved EcLeuRS-
pair that was derived from E. coli. Like 1, fluorescence of 2 is also sensitive to solvent polarity. Genetic incorporation of 2 into proteins can be applied to study protein folding/unfolding processes [65]. By genetically incorporating 2 into a voltage-dependent membrane lipid phosphatase, Wang and coworkers showed that 2 optically reports the conformational change of the voltage-sensitive domain in response to membrane depolarization [76]. Another fluorescent NAA that has been genetically incorporated into proteins is 3. Like 1 and 2, 3 is also an environment-sensitive fluorescent NAA. Using an evolved EcLeuRS- 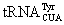 pair, 3 was successfully incorporated into glutamine-binding protein at the ligand binding site in yeast. The high fluorescent sensitivity of 3 in glutamine-binding protein to the polarity of the environment allowed easy detection of the conformation rearrangement of glutamine-binding protein during its strong association with glutamine [77].
pair, 3 was successfully incorporated into glutamine-binding protein at the ligand binding site in yeast. The high fluorescent sensitivity of 3 in glutamine-binding protein to the polarity of the environment allowed easy detection of the conformation rearrangement of glutamine-binding protein during its strong association with glutamine [77].
In comparison to protein-labeling approaches that need protein expression followed with additional chemical labeling, the direct incorporation of fluorescent NAAs into proteins is certainly a more optimal choice given its simplicity and efficiency. However, all three fluorescent NAAs that have been incorporated into proteins have relatively short fluorescent emission wavelengths. These narrow emission spectra and only a few available fluorophores limit the applications of the direct fluorescent NAA incorporation approach. Given the flexibility of PylRS and its variants in the recognition of different NAAs, other fluorescent NAAs could possibly be genetically encoded using evolved PylRS derivatives [78]. The orthogonal nature of the PylRS- 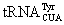 pair in bacteria, yeast, and mammalian cells will also make fluorescent NAAs encoded using this pair more applicable for broad applications. However, before additional fluorescent NAAs that cover a wide spectral range are genetically encoded, proteinlabeling approaches in which genetically encoded NAAs are chemically labeled with structurally diverse fluorophores are still necessary.
pair in bacteria, yeast, and mammalian cells will also make fluorescent NAAs encoded using this pair more applicable for broad applications. However, before additional fluorescent NAAs that cover a wide spectral range are genetically encoded, proteinlabeling approaches in which genetically encoded NAAs are chemically labeled with structurally diverse fluorophores are still necessary.
The concept of click reactions was introduced about a decade ago [79]. Reactions that can be classified as click reactions need to be selective, modular, and wide in scope. In order to undergo clicktype protein-labeling reactions, bioorthogonal functional groups that do not exist in the biological system can be introduced into proteins followed by selective reactions with fluorophore-containing dyes. One of these bioorthogonal functional groups is the keto group. Strictly speaking, keto is not totally bioorthogonal. It exists in the cellular metabolites, cofactors, and a small group of proteins [80,81]. However, in generally, keto is not found in proteins and DNA. Given that keto is the most versatile functional group in organic chemistry and selectively reacts with hydrazine- and hydroxylamine-containing molecules, its incorporation into proteins will make it possible to selectively label target proteins with hydrazine- and hydroxylamine-containing dyes. To genetically encode a keto functional group, Schultz and coworkers evolved several MjTyrRS variants for specific incorporation of a NAA 4 shown in Figure 2 into proteins in E. coli [62]. 4 has also been genetically encoded in yeast and mammalian cells using evolved tyrosyl-tRNA synthetase (EcTyrRS)-amber suppressing 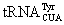 pairs that were derived from E. coli [53,54]. Brustad et al. demonstrated a grand application of 4 in the protein folding dynamic analysis [82]. Using orthogonal reactions with a genetically encoded 4 and a cysteine residue, two fluorescent dyes that formed a Förster Resonance Energy Transfer (FRET) pair were introduced in T4 lysozyme for single-molecule FRET analysis of protein folding. Another two NAAs that have been genetically incorporated into proteins using evolved MjTyRS variants are 5 and 6 [63,83]. By selectively targeting 5, Zhang et al. showed that a cytoplasmic Z domain protein and outer membrane protein LamB could be selectively visualized with a hydrazide-containing fluorescent dye [63]. Although 4-6 have been proved useful in selective protein labeling, their labeling efficiency at the physiological pH is low. Close to quantitative labeling with hydroxylamine-containing dyes could only be achieved at pH 4 with overnight incubation and labeling with hydrazide-containing dyes exhibited very low efficiency at pH 4-10 [82]. The low reactivity of the keto group in 4-6 is possibly due to its conjugation with an aromatic phenyl ring. The conjugative electrondonating effect of the phenyl group may reduce the electrophilicity of the keto carbonyl carbon and decrease its reactivity. To resolve this problem, Liu and coworkers designed another NAA 7 shown in Figure 2 [84]. The genetic incorporation of 7 was achieved using an evolved PylRS-
pairs that were derived from E. coli [53,54]. Brustad et al. demonstrated a grand application of 4 in the protein folding dynamic analysis [82]. Using orthogonal reactions with a genetically encoded 4 and a cysteine residue, two fluorescent dyes that formed a Förster Resonance Energy Transfer (FRET) pair were introduced in T4 lysozyme for single-molecule FRET analysis of protein folding. Another two NAAs that have been genetically incorporated into proteins using evolved MjTyRS variants are 5 and 6 [63,83]. By selectively targeting 5, Zhang et al. showed that a cytoplasmic Z domain protein and outer membrane protein LamB could be selectively visualized with a hydrazide-containing fluorescent dye [63]. Although 4-6 have been proved useful in selective protein labeling, their labeling efficiency at the physiological pH is low. Close to quantitative labeling with hydroxylamine-containing dyes could only be achieved at pH 4 with overnight incubation and labeling with hydrazide-containing dyes exhibited very low efficiency at pH 4-10 [82]. The low reactivity of the keto group in 4-6 is possibly due to its conjugation with an aromatic phenyl ring. The conjugative electrondonating effect of the phenyl group may reduce the electrophilicity of the keto carbonyl carbon and decrease its reactivity. To resolve this problem, Liu and coworkers designed another NAA 7 shown in Figure 2 [84]. The genetic incorporation of 7 was achieved using an evolved PylRS- 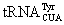 pair that was original used for the genetic incorporation of Nε-acetyl-lysine [85]. 7 contains an aliphatic keto group and is in theory more reactive toward hydrazine- and hydroxylamine containing dyes. As demonstrated, proteins incorporated with 7 could be quantitatively labeled with hydroxylamine-containing dyes or probes at the physiological conditions with 5 h incubation.
pair that was original used for the genetic incorporation of Nε-acetyl-lysine [85]. 7 contains an aliphatic keto group and is in theory more reactive toward hydrazine- and hydroxylamine containing dyes. As demonstrated, proteins incorporated with 7 could be quantitatively labeled with hydroxylamine-containing dyes or probes at the physiological conditions with 5 h incubation.
The copper catalyzed azide-alkyne Husigen cycloaddition (CuAAC) reaction is a typical click reaction [86]. Both azide and alkyne are biologically inert. Azide itself is also absent in the biological system and alkyne doesn’t exist in bio-macromolecules. Given the high reaction specificity and reactivity between azide and alkyne, specific installation of either azide or alkyne into a protein will confer labeling of this protein with a fluorescent dyes that contain a corresponding alkyne or azide functional group. The first NAA that was genetically incorporated into proteins for this purpose is 8 shown in Figure 3. The genetic incorporation of 8 into proteins was achieved in E. coli using evolved MjTyrRS- 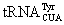 pairs and in yeast and mammalian cells using evolved EcTyrRS-
pairs and in yeast and mammalian cells using evolved EcTyrRS-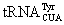 pairs [53,54,87,88]. Using the CuAAC reaction, proteins incorporated with 8 have been selectively labeled with alkyne-containing fluorescent dyes with high labeling efficiency. It was also demonstrated that the CuAAC reaction worked efficiently to fluorescently label phage particles incorporated with 8 [89]. Deiters et al. also demonstrated that the same reaction could be applied to PEGylate proteins incorporated with 8 [88].
pairs [53,54,87,88]. Using the CuAAC reaction, proteins incorporated with 8 have been selectively labeled with alkyne-containing fluorescent dyes with high labeling efficiency. It was also demonstrated that the CuAAC reaction worked efficiently to fluorescently label phage particles incorporated with 8 [89]. Deiters et al. also demonstrated that the same reaction could be applied to PEGylate proteins incorporated with 8 [88].
Although the CuAAC reaction has approved advantageous, the requirement to use Cu(I) as a catalyst does have some drawbacks. The Cu(I) catalyst can induce protein aggregation and oxidation, often obviating its application in living systems [90]. Phage particles incorporated with 8 were not viable when they were reacted with an alkyne-containing fluorescein dye in the presence of Cu(I) [91]. To improve biocompatibility of the CuAAC reaction, multiple new ligands of Cu(I) have been introduced [92,93]. One of these ligands not only largely shields the deleterious effects of Cu(I) and also increases its catalyzed reaction rate [92]. It has been applied in several studies for protein labeling in living cells [94]. There are also two alternative methods for chemically labeling azide-containing proteins without using catalysts. The Staudinger ligation reaction between an azide and a phosphine probe was developed by Bertozzi and coworkers and previously used to modify cell surface carbohydrates in both cellular and in vivo systems [95]. This reaction proceeds with excellent yields under the physiological conditions and is highly selective for azides. This reaction is also biocompatible. Phage particles incorporated with 8 were still viable after their reactions with phosphine-containing dyes [91]. The other reaction that can selectively label an azidecontaining protein without the use of a catalyst is the azide-cyclooctyne cycloaddition reaction [96]. This reaction was also developed by Bertozzi and coworkers. A cyclooctyne that has a strain-promoted alkyne functional group undergoes a rapid reaction with an azide. Using a cyclooctyne-containing dye, Liu and coworkers showed that proteins incorporated with 8 could be selectively and efficiently labeled [90].
Beside 8, two other azide-containing dyes 9 and 10 have also been incorporated into proteins in E. coli using evolved PylRS- 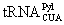 pairs [78,97]. Chen and coworkers showed that HdeA incorporated with 10 in E. coli could be selectively labeled with an alkyne-containing and environment-sensitive fluorescent dye. HdeA is an acid-resistant chaperon that shows pH-mediated conformational changes under low pH conditions. One HdeA variants that was fluorescently labeled showed a strong fluorescence increase upon acidification [98]. Since the PylRS-
pairs [78,97]. Chen and coworkers showed that HdeA incorporated with 10 in E. coli could be selectively labeled with an alkyne-containing and environment-sensitive fluorescent dye. HdeA is an acid-resistant chaperon that shows pH-mediated conformational changes under low pH conditions. One HdeA variants that was fluorescently labeled showed a strong fluorescence increase upon acidification [98]. Since the PylRS- 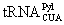 pair is orthogonal in yeast and mammalian cells, 9 and 10 could be potentially incorporated into proteins in these cellular systems for selective protein labeling.
pair is orthogonal in yeast and mammalian cells, 9 and 10 could be potentially incorporated into proteins in these cellular systems for selective protein labeling.
Alkyne-containing NAAs that have been genetically encoded are 11-16 in Figure 4. 11-13 contain a terminal alkyne. Proteins incorporated with these NAAs undergo the CuAAC reaction with azide-containing fluorescent dyes. Genetic encoding of 11 has been achieved in E. coli, yeast, and mammalian cells using evolved MjTyRS-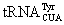 pairs, evolved EcTyrRS-
pairs, evolved EcTyrRS- 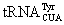 pairs, and a designed PylRS mutant-
pairs, and a designed PylRS mutant- 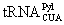 pair [54,99-101]. 12 and 13 are genetically encoded in cells using the wild type PylRS-
pair [54,99-101]. 12 and 13 are genetically encoded in cells using the wild type PylRS-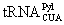 [78,102]. 14-16 contain a cyclooctyne moiety that undergoes the copper free azide-cyclooctyne cycloaddition reaction. All three NAAs have been genetically encoded in E. coli and mammalian cells using mutant PylRS-
[78,102]. 14-16 contain a cyclooctyne moiety that undergoes the copper free azide-cyclooctyne cycloaddition reaction. All three NAAs have been genetically encoded in E. coli and mammalian cells using mutant PylRS- 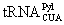 pairs [103,104]. Lemke et al. showed that a GFP variant mCherry incorporated with 14 in E. coli could be selectively lighted up with a coumarin azide.
pairs [103,104]. Lemke et al. showed that a GFP variant mCherry incorporated with 14 in E. coli could be selectively lighted up with a coumarin azide.
Besides the strain-promoted azide-cyclooctyne cycloaddition reaction, 14-16 also undergo strain-promoted inverse-electrondemanding cycloaddition with tetrazine-containing molecules that can exhibit accelerated reaction rates using strained reactants and furthermore is irreversible because of the loss of N2 [105]. This chemistry has been used in cells to label small molecules and is magnitudes faster that the classical CuAAC reaction. Lemke and coworkers showed that 14 and 15 efficiently reacted with a tetrazine-containing dye with a second order reaction rate reaching to 400 M-1s-1 [103,106]. Maltose binding protein incorporated with 14 in E. coli could be efficiently and rapidly labeled with a tetrazine-containing coumarin dye. In comparison to 14 and 15, 16 has a higher reaction rate with a tetrazine dye with a second order reaction rate close to 1200 M-1s-1 [104]. Fusion proteins such as EGFR-GFP and jun-mCherry incorporated with 15 in HeLa cells could be efficiently visualized with a TAMRA-tetrazine dye.
Other NAAs that undergo strain-promoted inverse-electrondemanding Diels-Alders cycloaddition with tetrazine-containing molecules include 17-19 in Figure 5. 17 and 18 contain a norbornene moiety that contains a strain-promoted alkene. The genetic incorporation of 17 and 18 has been achieved using mutant PylRS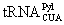 pairs [103,104]. Proteins incorporated with 17 and 18 reacted rapidly with tetrazine-containing dyes. Chin and coworkers showed that mammalian membrane proteins incorporated with 17 could be efficiently labeled with tetrazine-containing dyes [104]. 19 is also genetically encoded using mutant PylRS-
pairs [103,104]. Proteins incorporated with 17 and 18 reacted rapidly with tetrazine-containing dyes. Chin and coworkers showed that mammalian membrane proteins incorporated with 17 could be efficiently labeled with tetrazine-containing dyes [104]. 19 is also genetically encoded using mutant PylRS- 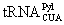 pairs [103,104]. It has a trans-cyclooctene moiety with a highly strain promoted alkene group. This strain-promoted alkene group can undergo inverse-electron demanding cycloaddition with a tetrazine dye with a reaction rate close to 35,000 M-1s-1. As far as we notice, this is the fastest click reaction that has been reported. A fusion protein, NLS-MBP-GFP incorporated with 19 in HeLa cells could be efficiently labeled with a tetrazine-containing Cy5 dye in just about 5 min incubation. This fast labeling process could effectively avoid exposing cells in non-native conditions and increase cell viability. There is another advantage of using inverse-electron-demanding Diels- Alder cycloaddition to label a strain-promoted alkene. The tetrazine moiety itself can efficiently quench fluorescence of a fluorophores that is covalently linked to it. Therefore, before its reaction with a strain promoted alkene-containing NAA, a tetrazine-containing dye is not fluorescent. However, the tetrazine moiety is lost after reaction and then the fluorophore emits strong fluorescence. Thus, a tetrazine containing dye is a “turn-on” fluorophore of a strain-promoted alkene. Cleaning the residual dye after reaction is not necessary given its low background.
pairs [103,104]. It has a trans-cyclooctene moiety with a highly strain promoted alkene group. This strain-promoted alkene group can undergo inverse-electron demanding cycloaddition with a tetrazine dye with a reaction rate close to 35,000 M-1s-1. As far as we notice, this is the fastest click reaction that has been reported. A fusion protein, NLS-MBP-GFP incorporated with 19 in HeLa cells could be efficiently labeled with a tetrazine-containing Cy5 dye in just about 5 min incubation. This fast labeling process could effectively avoid exposing cells in non-native conditions and increase cell viability. There is another advantage of using inverse-electron-demanding Diels- Alder cycloaddition to label a strain-promoted alkene. The tetrazine moiety itself can efficiently quench fluorescence of a fluorophores that is covalently linked to it. Therefore, before its reaction with a strain promoted alkene-containing NAA, a tetrazine-containing dye is not fluorescent. However, the tetrazine moiety is lost after reaction and then the fluorophore emits strong fluorescence. Thus, a tetrazine containing dye is a “turn-on” fluorophore of a strain-promoted alkene. Cleaning the residual dye after reaction is not necessary given its low background.
Like strain-promoted alkene-containing NAAs, a tetrazinecontaining NAA could also be genetically encoded. Using an evolved MjTyRS- 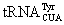 pair, 20 was genetically incorporated into proteins in E. coli [107]. Mehl and coworkers showed that 20 reacted with a strained trans-cyclooctene with a rate of 880 M-1s-1. Using this strained trans-cyclooctene in living E. coli cells, GFP incorporated with 20 that was not fluorescent due to the fluorescent quenching effect of 20 could be rapidly lighted up.
pair, 20 was genetically incorporated into proteins in E. coli [107]. Mehl and coworkers showed that 20 reacted with a strained trans-cyclooctene with a rate of 880 M-1s-1. Using this strained trans-cyclooctene in living E. coli cells, GFP incorporated with 20 that was not fluorescent due to the fluorescent quenching effect of 20 could be rapidly lighted up.
There are two other reaction types that may be considered as click reactions and used in selective protein labeling. Lin and coworkers showed that a tetrazole undergoes photolysis to form a nitrile imine that selectively reacts with an alkene [108]. This is called a photoclick reaction. Using a tetrazole-containing dye, Lin and coworkers demonstrated proteins incorporated with 21 (Figure 6) could be selectively labeled under UV irradiation [109,110]. A tetrazole-containing NAA, 22 was also genetically incorporated into proteins in E. coli [109]. Proteins incorporated with 22 underwent photo click reaction with alkene-containing dyes. Cyanobenzothiazole condensation with 1,2-aminothiol is another reaction type that is considered bioorthogonal. Chan, Chin, and coworkers showed that a 1,2-aminothiol-containing NAA 23 could be genetically incorporated into proteins in E. coli using either a wild type or evolved evolved PylRS-  pair [111,112]. The purified protein incorporated with 23 underwent efficient labeling with a cyanobenzothiazole containing dye.
pair [111,112]. The purified protein incorporated with 23 underwent efficient labeling with a cyanobenzothiazole containing dye.
So far, multiple bioorthogonal click type reactions have been developed for selective protein labeling. However, the orthogonal nature of these reactions to each other is not very much explored. Developing two orthogonal click reactions in living cells could be potentially important in selectively labeling one protein with two different dyes for protein folding/unfolding analysis inside living cells and selectively labeling two proteins with two dyes for their interaction analysis. Recently, Liu and Chin Groups independently developed two methods for genetic incorporation of two different NAAs into one protein in living cells [113,114]. These same systems could also be applied to synthesize two different proteins that are incorporated with two different NAAs for their following chemical modifications. In order to undergo selective modifications of two different NAAs in living cells, two orthogonal click reactions are necessary. We think one important future direction of the current NAA-directed fluorescent protein labeling research is to identify and optimize two orthogonal click reactions for rapid labeling of two different NAAs in one cell.
This work was partly supported by Welch Research Grant A-1715 to W. R. Liu.