Advancements in Genetic Engineering
Open Access
ISSN: 2169-0111
ISSN: 2169-0111
Research Article - (2015) Volume 4, Issue 2
The effect of oil-rich supplements on the expression of genes involved in lipogenesis and reproduction in pasturebased dairy cows is currently unknown, or at best, scanty and limited to impacts on cow liveweight, body condition score, milk composition, fatty acid and plasma metabolite profiles only. This research investigated the gene expression patterns of Aralkylamine N-acetyltransferase (AANAT), B-cell translocation gene-2 (BTG2) and Fatty Acid Synthase (FASN) genes in response to incremental levels of dietary crude degummed canola oil (CDCO). We tested the hypothesis that the relative mRNA abundance and gene expression profiles of AANAT, BTG2 and FASN in primiparous Holstein-Friesian cows will be up-regulated in response to post-partum dietary supplementation with CDCO in a typical pasture-based dairy production system. Thus, the primary objective of this study was to investigate the expression of AANAT, BTG2 and FASN genes in response to incremental levels of CDCO. A random allocation of primiparous Holstein-Friesian dairy cows into four treatment groups comprising wheat-based pelleted with no supplemental CDCO (control), or with CDCO added at 25 ml kg-1 DM (low), 35 ml kg-1 DM (medium) and 50 ml kg-1 DM (high) was utilized in a ten-week experimental feeding trial including two weeks of adjustment. Both level of supplementation and their interaction with duration were significant sources of variation (P<0.05) that influenced BTG2 expression, while the expressions of AANAT and FASN genes were unaffected (P>0.05). The high (0.67 fold), medium (0.87 fold) and low (0.56 fold) treatments had suppressed BTG2 expressions compared to the control (1.0 fold) group. The low expression of BTG2 might be important when the reproductive system of cows is recovering from the effect of gestation and new cell growth is required.
<Keywords: Gene expression; mRNA; Canola oil; Aralkylamine Nacetyltransferase; B-cell translocation gene-2; Fatty acid synthase; Pasture-based Holstein-Friesian cows
AANAT: Aralkylamine N-acetyltransferase; ADF: Acid Detergent Fibre; APRO: Anti-Proliferative; CDCO: Crude Degummed Canola Oil; AI: Artificial Insemination; BCS: Body Condition Score; BLAST: Basic Local Alignment Search Tool; BTG2: B-cell Translocation Gene 2; cDNA: Complementary Deoxyribonucleic Acid; CLA: Conjugated Linoleic Acid; CP: Crude Protein; Ct: Cycle threshold; DM: Dry Matter; FASN: Fatty Acid Synthase; GLC: Gas Liquid Chromatography; "LC ω-3" to "LC ω-3": Long chain Omega-3; ME: Metabolisable Energy; NADPH: Nicotinamide Adenine Dinucleotide Phosphate; NDF: Neutral Detergent; PUFA: Polyunsaturated Fatty Acids; qPCR: Quantitative Real Time Polymerase Chain Reqaction; RNA: Ribonucleic Acid; SAS: Statistical Analysis System; SFA: Saturated Fatty Acids; TIA: Tasmanian Institute of Agriculture
Nutritional attempts to remedy infertility are of interest to the dairy industry [1] because the antagonistic relationship between high milk production and fertility in modern, high genetic-merit cows has concomitantly led to a gradual but progressive decline in reproductive performance in diverse dairy production systems around the world. Prolonged calving intervals along with embryonic losses and postpartum anovulatory intervals are some of the major causes of infertility in cows [2]. In a typical pasture-based dairy system, different sources of lipids fed to lactating cows have been trialled to primarily increase the energy density of the diet in order to enhance milk production when negative energy balance peaks [3]. Research findings suggest that dietary supplementation with fat sources containing adequate proportions of unsaturated fats could potentially improve fertility in high merit dairy cows [4]. Therefore, a new, effective and long-term nutritional strategy that can assist in a better understanding of nutrition-fertility interactions in pasture-based systems is a potential solution to the subfertility problem in dairy cows.
Lipids also epitomize an effective nutritional approach for modifying milk fat composition [5] to favour an elevated profile of beneficial polyunsaturated fatty acids [6]. Lipids also play a crucial role in regulating the expression of genes essential for fertility and de novo fat synthesis in dairy cows. It has been shown that dietary fats containing trans-10, cis-12 conjugated linoleic acid (CLA) cause milk fat depression by inhibiting the expression of fatty acid synthase (FASN) gene [3] which is known to play a central role in the biosynthesis of fat in the mammary gland of mammals [7]. Although FASN is an important gene involved in lipogenesis, there is only limited published information about its expression in fatsupplemented cows in a pasture-based dairy production system. Where such studies were conducted, results have been conflicting and inconsistent [8] and warrant further research.
Arylalkylamine N-acetyltransferase (AANAT) is an essential gene for melatonin biosynthesis [9]. Melatonin is directly associated with optimal functioning of the ovary, where it regulates the hypothalamicpituitary- gonadal axis to instigate folliculogenesis and steroidogenesis [10,11]. However, previous studies on AANAT have focused mainly on humans, inspite of AANAT being an important gene controlling reproduction in other mammals [12]. Currently, there is limited information on the expression of AANAT gene in dairy cows, especially with regards to fat supplementation, thus creating an important knowledge gap that this study intends to fill.
B-cell translocation gene-2 (BTG2) is an anti-proliferative gene that regulates cell cycle growth and BTG2 research investigations have been limited to cancer studies [13] demonstrating that the anti-proliferative characteristic of BTG2 gene is crucial during ovulation in mammals [14]. Published information on the expression and function of the BTG2 gene in dairy cattle, especially when supplemented with dietary lipids in a pasture-based system, are to our knowledge, either nonexistent or at best, scanty. The above mentioned genes are related with differences in total fatty acid content in animal tissues and the protection of long chain (≥C20) polyunsaturated fatty acids through the prevention of peroxidation [15].
Understanding the mechanism underpinning the impact of dietary fat intake on the reproductive sequences from oestrous to conception in cows could revolutionise how nutrition is managed in dairy farms to improve reproductive performance. It will also assist researchers in unravelling the mystery behind the current global and gradual, but progressive, decline in dairy cow fertility. In this regard, further studies are required to unpack the intricate biological mechanisms involved with feeding dietary fats to grazing cows and their effects on lactation and fertility traits. This will enable dairy farmers make informed choices and tailored decisions when feeding lactating cows with specific dietary fat supplements. We hypothesized in this study that supplementation with CDCO would affect the expression of genes involved in reproductive functions (AANAT, BTG2) and de novo fatty acid synthesis (FASN) in primiparous Holstein-Frisian cows grazing under similar environmental conditions. Therefore, the primary objective of this study was to determine the relative abundance and expression of genes encoding proteins required for optimal reproduction and de novo lipogenesis in pasture-based lactating cows subjected to zero, low, medium and high levels of dietary supplementation with CDCO.
Animal ethics
The use of animals and procedures performed in this study were all approved by the University of Tasmania Animal Ethics Committee (Permit No AA0012583), and were conducted in accordance with the 1993 Tasmanian Animal Welfare Act and the 2004 Australian Code of Practice for the Care and Use of Animals for Scientific Purposes.
Experimental site and location
The experiment was carried out at the University of Tasmania’s Dairy Research Centre, Tasmanian Institute of Agriculture (TIA) Elliot Dairy Research Farm in Somerset, North-Western Tasmania, Australia, from September to November 2012. Tasmania is Australia’s smallest state with a land size of 68,000 square kilometers and located within the cool, temperate, climatic zone at latitude 42° South and longitude 147° East. It is characterized by four distinct seasons - winter, autumn, spring and summer. The experiment was carried out in spring when the annual rainfall was 2500 mm and humidity was approximately 60%.
Experimental design, treatment groups and supplementary feeding trial
The physical condition and energy status of the experimental cows was visually assessed based on body condition score (BCS) on a scale of 1-8 [16]. Twenty primiparous, spring-calving, purebred, Holstein- Friesian cows (average liveweight of 400 ± 40 kg, BCS 4 ± 1.0 and 40 ± 8 days in milk (DIM), were randomly allocated into 1 of 4 treatment groups of supplementation with wheat-based CDCO pellets classified as low (25 ml/ kgDM), medium (35 ml/ kgDM), high (50 ml/ kgDM) and the control (no CDCO- 0 ml/ kgDM). This replicated herd of cows (n=5 per treatment group) receiving CDCO supplements was placed under the same management and rotated in electric fenced paddocks with the Control cows offered wheat-based pellets without CDCO. Together, the animals had access to 3000 kgDM of forages, a mixture of ryegrass (Lolium perenne), cocksfoot (Dactylis glomerata), and white clover (Trifolium repens) pasture grazed at the two-leaf stage. Water was offered ad libitum. The current level of CDCO was calculated based on 7% total fat allowed in the diet of grazing cows. Each cow received 6 kg of the pelleted supplements daily for eight weeks, after two weeks of adjustment. Supplements were offered to cows in two splits; morning (3 kg) and evening (3 kg) milking sessions at 0500 hr and 1500 hr. There was no feed residual left over from any of the groups. The exact pasture intake was difficult to estimate as the case is under grazing conditions.
Feed chemical composition analysis
The physical condition Dry matter (DM) content of the basal and experimental diets was determined by drying samples to a constant temperature at 65°C in a fan forced oven, finely ground to pass through a 2 mm sieve using Laboratory Mill (Thomas Model 4 Wiley® Mill; Thomas Scientific), and further drying at 105°C for 24 h. The DM was computed as the difference between the initial and final weights of samples. Ash content was determined by combusting samples in a furnace at 600°C for 8 hours. Neutral detergent (NDF) and acid detergent fibre (ADF) contents were measured using an Ankom Fiber Analyzer (ANKOM220; ANKOM Technology, USA). Nitrogen content was determined using a Thermo Finnigan EA 1112 Series Flash Elemental Analyzer and the values multiplied by 6.25 to give the crude protein (CP) percentage. Ether extract (EE) was determined using an Ankom fat/oil extractor (ANKOMXT15; ANKOM Technology, USA). Metabolisable energy (ME) was calculated in accordance with Van Es [17]. The chemical compositions of the treatment, control and basal feeds are presented in Table 1.
| a-bChemical composition (%DM) | cFeeds | ||
|---|---|---|---|
| Treatment (canola oil) | Control(No canola oil) | Basal diet(Pasture) | |
| MC | 8.2 | 9.1 | 5.5 |
| DM | 91.8 | 90.9 | 94.5 |
| ADF | 8.0 | 9.0 | 27.7 |
| NDF | 20.0 | 21.1 | 45.9 |
| EE | 6.2 | 2.1 | 3.0 |
| Ash | 9.7 | 8.9 | 9.3 |
| NFC | 52.8 | 59.0 | 23.9 |
| OM | 90.3 | 91.1 | 90.7 |
| CP | 12.7 | 10.4 | 21.0 |
| ME (MJ/kg DM) | 4.08 | 4.07 | 3.99 |
aAll feeds were analysed based on a dry weight basis; bMoisture content (MC), Dry matter (DM), organic matter (OM), neutral detergent fibre (NDF), acid detergent fibre (ADF), non-fibrous carbohydrate (NFC), ether extract (EE), crude protein (CP) and Metabolisable energy (ME). Treatment: feed with added canola oil. c Control: feed without canola oil, Basal diet: mixed ryegrass pasture.
Table 1: Chemical composition of the experimental and basal feeds.
Fatty acid analysis of basal and supplementary feeds
The fatty acid profiles of both basal and supplementary feeds were analysed by gas liquid chromatography (GLC) and presented in Table 2. The detailed procedure had been previously described and published [18].
| aFatty acid | bFeed components | ||
|---|---|---|---|
| Control(No canola oil)% | Treatment (canola oil)% | Basal (Pasture)% | |
| 12:0 | 0.00 | 0.00 | 0.05 |
| 14:0 | 0.10 | 0.09 | 0.10 |
| 15:0 | 0.20 | 0.13 | 0.20 |
| 16:1 | 0.00 | 0.00 | 1.00 |
| 16:0 | 32.10 | 26.10 | 10.00 |
| 17:0 | 0.20 | 0.18 | 0.10 |
| 18:3ω 6 | 0.00 | 0.03 | 0.00 |
| 18:4ω 3 | 0.00 | 0.00 | 0.90 |
| 18:2ω 6 LA | 17.70 | 6.86 | 9.10 |
| 18:3ω 3 ALA | 1.60 | 0.48 | 64.30 |
| 18:1ω 9c | 16.50 | 41.90 | 4.40 |
| 18:1ω 7t | 0.20 | 0.10 | 0.20 |
| 18:0 | 3.80 | 3.83 | 2.20 |
| 18:2CLA | 0.10 | 1.48 | 0.00 |
| 19:0 | 0.90 | 3.47 | 0.10 |
| 20:4ω 6 | 0.00 | 0.01 | 0.00 |
| 20:5ω 3 EPA | 11.80 | 0.20 | 0.10 |
| 20:3ω 6 | 0.40 | 1.82 | 0.80 |
| 20:4ω 3 ARA | 0.40 | 0.22 | 0.10 |
| 20:2ω 6 | 1.40 | 1.45 | 0.00 |
| 20:0 | 0.80 | 1.38 | 0.40 |
| 22:5ω 6 | 0.30 | 0.04 | 0.10 |
| 22:6ω 3 DHA | 0.20 | 0.03 | 0.00 |
| 22:4ω 6 | 0.20 | 0.00 | 0.00 |
| 22:5ω 3 DPA | 0.90 | 0.00 | 0.00 |
| 22:0 | 1.80 | 1.86 | 1.50 |
| 24:0 | 1.10 | 1.30 | 0.90 |
| tSFA | 41.20 | 38.64 | 16.45 |
| tMUFA | 23.30 | 48.74 | 8.00 |
| tPUFA | 35.00 | 12.62 | 75.40 |
| ω-3 PUFA | 14.90 | 0.93 | 65.40 |
| ω-6 PUFA | 20.10 | 10.24 | 10.10 |
| ω-3 LC-PUFA | 13.30 | 0.45 | 0.20 |
aεtSFA is the sum of 12:0, 13:0, i14:0,14:0, i15:0, a15:0,15:0, i16:0, 16:0, i17:0, 17:0, i18:0, 18:0, 19:0,20:0, 20:0, 22:0, 24:0; εtMUFA is the sum of 14:1ω -5c, 15:1ω-6c, 16:1ω -9c, 16:1ω -7c, 16:1ω -7t, 16:1ω -5c, 16:1,17:1ω -8+a17:0, 17:1ω-6c, 18:1ω -9c, 18:1ω -7c, 18:1ω -7t, 18:1ω -5c, 18:1a, 18:1b , 20:1ω -11c, 20:1ω -9c, 20:1ω -7c, 20:1ω -5c, 22:1ω -11c, 22:1ω -9c, 22:1ω -7c, 24:1ω -11c, 24:1ω -9c, 24:1ω -7c; εtPUFA is the sum of 18:3ω-6, 18:4ω-3, 18:2ω-6, 18:3ω-3, 18:2CLA, 20:4ω-6, 20:5ω-3, 20:3ω-6, 20:4ω-3, 20:2ω-6, 22:5ω-6, 22:6ω-3, 22:4ω-6, 22:5ω-3; εω-3 LC-PUFA is the sum of 20:5ω-3, 20:4ω-3, 22:6ω-3, 22:5ω-3; εω-3 PUFA is the sum of 18:4ω-3, 18:3n-3, 20:4ω-3, 20:5ω-3,22:6ω-3, 22:5ω-3; εω-6is the sum of 15:1ω-6, 17:1ω-6, 18:2ω-6, 18:3ω-6, 20:4ω-6, 20:3ω-6, 20:2ω-6, 22:5ω-6, 22:4ω-6. tSFA= total saturated fatty acids,tMUFA: total Monounsaturated Fatty Acids, tPUFA:Total Polyunsaturated Fatty Acids, ω-3 FA:Total Omega-3 Fatty Acids, ω-6 FA: Total Omega-6 Fatty acids, ω-3 LC-FA: Total Omega-3 Long Chain Fatty Acids; bControl:feqed with no Added Canola Oil; Treatment:Feed with Canola Added; basal:Mixed Ryegrass Pasture.
Table 2: Fatty acid concentration as a percentage of total fatty acids of basal and supplementary feeds.
Blood sample collection
Blood samples were collected from all experimental cows by coccygeal venipuncture into vacutainers containing heparin after the morning milking (0500 hr) on the day before the initiation of supplementation with CDCO and in week eight at the conclusion of the experiment. More frequent blood sample collection interval was restricted by the terms and conditions of the Animal Ethics Permit No AA0012583 granted by the University of Tasmania Animal Ethics Committee. The samples were immediately frozen in -20°C and transported to the laboratory for further storage in -80°C until RNA extraction.
RNA extraction and cDNA synthesis
Frozen blood samples were thawed and utilised for the isolation of total RNA using TRIzol® Plus RNA Purification Kit (Life Technologies Pty Ltd. Victoria, Australia). A tissue lyser (Qiagen Ltd., Crawley, UK) was used to homogenise the sample in TRIzol® Reagent. Total RNA quantity and quality was measured using the NanoDrop 8000 spectrophotometer (NanoDrop, Wilmington, DE, USA). RNA that had an absorbance (A260/280) reading between 1.8 and 2 was deemed of good quality. The RNA samples were treated with PureLinkTMDNase (Life Technologies Pty Ltd. Victoria, Australia) and purified using the RNeasy1 Mini Kit (Qiagen Ltd, NSW, Australia). DNase-treated and purified total RNA was then reverse transcribed to cDNA with Mixed Oligo dT/Random Hexamer Primers using the Tetro cDNA Synthesis Kit (Bioline Pty Ltd. NSW, Australia) according to the manufacturer’s instructions.
Primer design and reference gene selection
All candidate and reference gene primers (Table 3) were designed using the Primer3 web-based software program (http:// frodo.wi.mit.edu/primer3/) from GeneWorks Pty Ltd, SA, Australia). Basic Local Alignment Search Tool (BLAST) from the National Centre for Biotechnology Information (http://www.ncbi.nlm.nih.gov/ BLAST/) was used to check for the specificity of the primers. The validity of all primers was confirmed using a serial dilution of pooled cDNA to create a standard curve. Subsequently, the amplified PCR products were sequenced to confirm their primer specific identity (Beckman Coulter CEQTM 8000 Series Genetic Analysis System). The mRNA abundance was determined using highly stable reference genes. The normalisation of expression data for the target genes Aralkylamine N-acetyltransferase (AANAT), B-cell translocation gene 2 (BTG2), and Fatty acid synthase (FASN) utilised two reference genes, Ubiquitin C (UBC) and Peptidyl-prolyl cis-trans isomeraseA (PPIA). A good selection criterion of reference genes was an expression ratio that was constant across all samples. The software program geNorm, version 3.5 [19], was used to calculate, confirm and validate the expression stability (M-value) of the reference genes.
| aGene symbol | qPCR Primers | bTa | Amplicon Size (bp) | |
|---|---|---|---|---|
| Forward Primer | Reverse Primer | |||
| AANAT | ACTGACCTTCACGGAGATGC | TTCACTCATTCTCCCCGTTC | 60 | 211 |
| ADRB3 | TCAGTAGGAAGCGGGTCGGG | GGCTGGGGAAGGGCAGAGTT | 60 | 291 |
| BTG2 | CTGGAGGAGAACTGGCTGTC | AAAACAATGCCCAAGGTCTG | 60 | 194 |
| FASN | GTGTGGTACAGCCCCTCAAG | ACGCACCTGAATGACCACTT | 60 | 110 |
| Reference genes | ||||
| UBC | CGTCTTAGGGGTGGCTGTTA | AAATTGGGGTAAATGGCTAGA | 60 | 90 |
| PPIA | TCATTTGCACTGCCAAGACTG | TCATGCCCTCTTTCACTTTGC | 60 | 72 |
aAralkylamine N-acetyltransferase: AANAT, β3-adrenergic receptor: ADRB3, B-cell translocation gene 2: BTG2, Fatty acid synthase: FASN, Ubiquitin C: UBC, Peptidyl-prolyl cis-trans isomeraseA: PPIA, bTa: Empirical annealing Temperature.
Table 3: Primer pairs designed for real-time PCR (qPCR).
Quantitative real time PCR (qPCR)
Following reverse transcription, cDNA quantity was determined and standardised to the required concentration for qPCR. Triplicate 20 μL reactions were carried out in a 72-well Rotor-Gene (QIAGEN GmbH, Hilden, Germany), containing 4 μL cDNA (50 ng), 10 μL 2× SensiFAST SYBR No-ROX Mix (Bioline Pty Ltd., NSW, Australia), 4.4 μL DEPC H2O, and 0.8 μL forward and reverse primers (100 fmol). Assays were performed using the Rotor-Gene 3000 (QIAGEN Pty Ltd., VIC, Australia) with the following cycling parameters: 95°C for 2 min polymerase activation; 40 cycles of 95°C for 5 s denaturation, 60°C for 10 s annealing and 72°C for 5 s extension. Gene expression levels were recorded as Ct values (i.e., the number of PCR cycles at which the fluorescence signal was detected above the threshold value) and all samples were run in triplicates. Amplification efficiencies were determined for all candidate and reference genes using the formula E: 10^(−1/slope), with the slope of the linear curve of cycle threshold (Ct) values plotted against the log dilution as per Higuchi et al., [20]. Primer concentrations were optimised for each gene and disassociation curves were examined for the presence of a single PCR product. The efficiency of the reaction was calculated using a 5-fold serial dilution of cDNA and generation of a standard curve. All PCR efficiency coefficients were between 1.7 and 1.8 and therefore deemed acceptable. The Rotor-Gene 3000 (version 6.0.16) (QIAGEN Pty Ltd., VIC, Australia) was used for efficiency correction of the raw Ct values. This process involved an inter-plate calibration based on a calibrator sample included on all plates, averaging of replicates, normalisation to the reference gene and the calculation of quantities relative to the highest Ct and log2 transformation of the expression values for all genes. A PCR efficiency coefficient between 1.7 and 1.8 was considered adequate. The mathematic model used to determine the expression level of the target gene in comparison to the reference gene is given below as per Pfaffl [21].
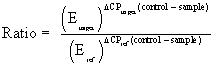
Statistical analysis
Initially, summary statistics by level and week (duration) of CDCO supplementation were computed to give means, standard deviations, standard error, variance, minimum and maximum values that were scrutinised for any data entry errors. Testing for linear, cubic and quadratic orthogonal contrasts by regressing the dependent on explanatory variables was carried out using PROC REG (SAS 2009) [22]. However, the linear, quadratic and cubic orthogonal contrasts were all found to be inconsequential. Therefore, repeated measures analysis of variance using PROC MIXED (SAS 2009) [22] was employed fitting fixed effects of treatment, week of supplementation and their second-order interactions on the expressions of AANAT, BTG2 and FASN genes. The 1st-order autoregressive covariance structure and level of supplementation were fitted as the repeated effects and cows as random effects. The degrees of freedom were estimated by the Satterthwaite method (SAS, 2009) [22]. Variable means are presented in Tables and Figures as LSM ± SEM. Tukey’s pairwise comparison test was utilized in establishing differences between means using the P<0.05 threshold for significance unless otherwise stated.
The effects of supplementation with CDCO on milk yield, milk fat and number of services per conception are presented in Figures 1-3, respectively. Cows receiving 50 mL kg-1 DM (High) of canola oil produced more milk (168.7 ± 3.4 vs. 157.1 ± 3.7 Litres) with a lower fat percentage (3.3 ± 0.1 vs. 4.0 ± 0.2%) than unsupplemented cows in the control treatment group (0 mL kg-1 DM) as depicted in Figures 1-3.
Figures 4-6 and Table 4 represent the relative mRNA abundance and expressions of AANAT, BTG2 and FASN genes, and these are individually highlighted as follows.
Figure 5: Effect of supplementation with CDCO on mRNA expression of the BTG2 gene in grazing cows (Mean ± SEM).
Aralkylamine N-acetyltransferase (AANAT)
Dietary supplementation of primiparous Holstein-Friesian cows with CDCO had no effect (P>0.05) on the expression of AANAT gene (Figure 4). As the week (duration) of supplementation progressed, the impact of CDCO supplements and interaction with duration of supplementation was insignificant on the expression of AANAT gene (Table 4).
| a Effect | b Genes | ||
| AANAT | BTG2 | FASN | |
| c TRT | 0.2019 | 0.0495 | 0.9289 |
| d Week | 0.2713 | 0.1818 | 0.3314 |
| e Week*TRT | 0.4956 | 0.0107 | 0.6647 |
aAll p-values in bold were significant (P<0.05). b Arylalkylamine-N-acetyltransferase (AANAT), B-cell translocation gene-2 ( BTG2), Fatty acid synthase(FASN), c TRT, treatment, d Week, week of lactation, e Week*TRT, interaction between week of supplementation and treatment.
Table 4: Multi-trait analysis of variance (p-values) for fixed and interaction effects of treatment and week of supplementation on the relative mRNA expression of AANAT: Arylalkylamine-N-acetyltransferase; BTG2: B-cell Translocation Gene-2; FASN: Fatty Acid Synthase genes in grazing Holstein-Friesian dairy.
B-cell translocation gene 2 (BTG2)
It was evident that both treatment and treatment by week of supplementation interactions were significant sources of variation that influenced the expression of BTG2 gene (P<0.05; Table 4). However, week of supplementation alone had no influence (P>0.05) on BTG2 gene expression. Cows receiving dietary supplementation with CDCO experienced consistent suppression of BTG2 gene expression compared to the control group (Figure 5). The cows in the control group recorded the greatest mRNA abundance of BTG2 (1.00 fold), followed by the medium group (0.87 fold), then the high group (0.67 fold) and finally the low group (0.56 fold).
Fatty acid synthase (FASN)
Differences in CDCO content in the supplemented primiparous Holstein-Frisian cows had no significant effect on the relative mRNA abundance of FASN gene. Week of supplementation and week by treatment interaction were inconsequential sources of variation (P>0.05).
A complex regulatory network of lipogenic genes, genetic and dietary differences impact the utilisation of oil-rich supplements in ruminants. To our best knowledge, the molecular mechanisms underlying these systems remain to be fully understood and characterized in pasture-based dairy production systems. Therefore, an attempt at trying to fathom the factors affecting dietary lipid metabolism in lactating cows is of utmost scientific relevance. This study utilised an experimental feeding trial with oil-rich dietary supplementation with CDCO in grazing primiparous Holstein- Friesian cows and determined the mRNA expression levels of BTG2, AANAT and FASN genes
BTG2
The mammalian BTG2 gene belongs to the anti-proliferative (APRO) family of genes that regulate cell cycle progression in a variety of cell types [23,24]. BTG2 is a prototypical member of the BTG/TOB family with anti-proliferative properties. The protein encoded by this gene controls cell cycle progression and proneural gene expression by acting as a transcription co-regulator that enhances or inhibits the activity of transcription factors [23,24]. Thus, BTG2 has many functions involving regulation of cell growth, death, differentiation and survival [25].
The present study found that supplementation of lactating Holstein-Friesian cows with CDCO repressed the expression of BTG2 significantly. The current result lends credence to the report of Jeckel et al. [26] who demonstrated that in rats, BTG2 gene was downregulated when dietary fatty acid was fed. Therefore, lack of BTG2 upregulation in the in the blood of cows in the present study would be due to the fatty acid content of the fed diet. Although some of the BTG2 gene expression studies were on tumorous diseases in humans, significant expressions have been observed in pig muscle, uterus and heart where the gene appears to play a role in cell development [27]. A previous study also found that gonadotropin hormones stimulate the expression of BTG2 genes in the ovary during ovulation [28]. Usually, luteinising hormone surges during the pre-ovulatory period, stops further growth of immature follicles, culminating in the ovulation of matured follicles for fertilisation [14,29]. The BTG2 gene is thus an essential gene for normal reproduction in mammals. The greater number of services per conception observed in the CDCO supplemented group suggests that these cows ovulated later than the control cows (Figure 3). This statement is supported by the low expression of BTG2 in supplemented cows (Figure 5). A previous study found that BTG2 gene acts as a co-activator for regulating hepatic gluconeogenesis [30]. In other words, the overexpression of BTG2 gene triggers hepatic gluconeogenesis, thus generating a spike in glucose synthesis which provides surplus energy for cell proliferation. It is well known that optimal recovery of the cow’s uterus following a normal gestation period requires growth, development and maturation of the granulosa/follicular cells for the cow to be ready for the next cycle of gestation. However, the anti-proliferative activity of the BTG2 gene might impact negatively on cell growth and tissue repair of the reproductive machinery. Our current findings seem to suggest that supplementation of cows with lipid-rich feeds could be utilised as a dietary manipulation tool to repress the expression of BTG2 gene and its anti-proliferative attributes.
AANAT
From published literature, the AANAT gene has been reported to be associated with long-chain omega-3 (LC ω-3) polyunsaturated fatty acid (PUFA) synthesis [15]. A major cause for disparity between this report and our observation in the current study could be due to differences in dietary lipid sources, dosages fed and the relatively smaller proportion of LC ω-3 PUFA in our experimental diet compared to the control diet (0.45% vs. 13.30%; Table 2). The AANAT gene is also known to encode an acetyltransferase superfamily protein [31] that catalyses the rate-limiting step in the synthesis of melatonin from serotonin [9] primarily found in the pineal gland [32]. Melatonin (N-acetyl-5-methoxytryptamine) is a hormone that plays a role in reproductive functions in mammals [33], particularly in the growth and maturation of oocytes in the ovary and steroidogenesis in the granulosa cells via the mitogen-activated protein kinase pathway [10,34]. Melatonin is also essential for the function of the circadian clock that influences activity and sleep [31,35]. The mechanism by which melatonin regulates reproduction has been reported to be through the control of gonadotropin releasing hormone (GnRH) and gonadotropin inhibiting hormone (GnIH) receptors primarily found in the hypothalamic-pituitary axis to release gonadotropin hormones [11,33,36]. AANAT transcripts have been found to be differentially expressed in high vs. low omega-3 index (O3I) muscles, suggesting a role for melatonin in reducing oxidative damage, including that to PUFA [9]. The ability of melatonin to protect against lipid peroxidation has been repeatedly documented in many studies using animal and plant tissues [37]. Spanish scientists reported that melatonin consumption assists in the control of weight gain since it stimulates the appearance of brown fat (beige), a type of fat cell that burns calories instead of storing them [38]. Their research demonstrated that melatonin treatment not only induced browning of inguinal white adipose tissue in Zucker diabetic fatty rats, but also increased thermogenic activity [38]. Taken together, these findings highlight the anti-obesity effect of melatonin and explain its metabolic benefits of protecting against oxidative degradation of PUFA in the muscle tissue thereby producing higher O3I levels [38].
The afore-mentioned body of evidence in the published literature indicates that the expression of AANAT gene could play multifaceted functions in regulating fertility in dairy cows through biosynthesis of melatonin. This makes AANAT an ideal gene to explore in terms possible nutritional manipulation of its expression to assist in controlling seasonal breeding in pasture-based dairy systems. From the current study, the supplementation of Holstein-Friesian cows with CDCO was inconsequential to AANAT gene expression since no differential expression of the AANAT gene was observed between the treatment groups. This suggests that supplementing grazing primiparous cows with CDCO may not be an influential determinant of AANAT gene expression. However, further research studies are warranted with different dietary fat sources to establish ideal dosage levels that can strongly up-regulate/and or down-regulate AANAT in dairy cows before its use can be adopted by the industry.
FASN
FASN encodes a multifunctional enzyme that catalyses fatty acid synthesis [39]. FASN is considered as a fundamental enzyme in de novo lipogenesis in mammals and its main function is to catalyse the synthesis of palmitate from acetyl-CoA and malonyl-CoA, in the presence of NADPH, into long-chain saturated fatty acids (LC-SFA) [39,40]. It has been shown that the FASN gene contributes to the regulation of body weight in humans, which results in the development of obesity [39,41]. Thus, FASN is a complex homodimeric gene that plays a pivotal role in de novo lipid biosynthesis and regulation of milk fat content [7]. The lack of significant effect of the experimental lipid-rich diet on FASN expression is in agreement with the report of Bichi et al. [42]. It is pertinent to state that an observed trend in the current study towards the suppression of FASN expression (Figure 6) agrees with previous research in cattle [32,43,44] and ewes [45].
The observed suppression trend of FASN expression is consistent with the role of fats containing conjugated linoleic acid (CLA), especially trans-10, cis-12 18:2 [34]. The fatty acid profile of our treatment diet had a greater proportion of CLA than the control diet (1.48% versus 0.10%). This could help to explain the down-regulation of FASN expression in cows supplemented with CDCO and also the suppression of the milk fat content (Figure 2). However, the discrepancy between the present and previous studies could be due to differences in the dietary fat source, dosage, level of polyunsaturated fatty acid intake, rumen biohydrogenation intermediates, and basal diet offered. One clear message from our results is the potential for supplementation of grazing cows with CDCO to down-regulate FASN expression; that way, the energy spared from reduced milk fat synthesis could be partitioned towards metabolic functions in nonmammary tissues, specifically, in reproductive tissues postpartum or milk production (Figure 1). This could prove highly significant to pasture-based dairy farmers in terms of managing the energy needs for production and reproduction in their herds. The downside to this assertion in pasture-based dairy systems is that milk fat is an economically important constituent of total milk solids upon which Tasmanian dairy farmers are paid in Australia.
Furthermore, extreme suppression of FASN expression may be adverse to the butter manufacturing industries due to lower milk fat content. However, the beneficial impact is that the milk is being rid of mostly saturated fats, thus giving consumers a healthier product for which a premium can be charged to compensate for lower milk solids.
Dietary supplementation of grazing primiparous Holstein-Frisian dairy cows with CDCO had a significant influence in down-regulating the expression of BTG2. This might be important when the reproductive system of cows is recovering from the effect of gestation and new cell growth is required, but the downside is that hepatic gluconeogenesis could be hampered. mRNA abundance and expression of AANAT and FASN genes were not significantly impacted by CDCO supplementation inspite of the observed trends towards up-regulation and down-regulation, respectively. The observed trend toward the suppression of FASN gene expression can be beneficial in sparing energy from milk fat synthesis and re-directing the surplus to non-mammary tissues in vivo. However, severe milk fat depression may be economically undesirable to Tasmanian dairy farmers because of its contribution to total milk solids upon which milk prices are based. These findings highlight the important role of nutrition in altering reproductive and lipogenic genes in the blood of lactating cows. Further studies with more experimental animals and CDCO supplementation levels would be required to confirm the current findings.
The authors declare that they have no competing interests.
This work was carried out in collaboration between all authors. Author JRO ran the feeding trial, laboratory analyses, collected experimental samples, performed feed chemical analysis and wrote the first draft of the manuscript as part of his PhD project. Authors JRO, BS, AK designed the primers, selected reference genes, extracted RNA, synthesised cDNA, and carried out quantitative real time PCR. Authors BSMA, PDN and AEOMA contributed in the reading and making needed changes to the draft manuscript. Author AEOMA conceived the research idea, wrote the funding grant, experimental design, read and made final changes to the final manuscript as a research article. All authors read and approved the final manuscript.
This study was supported with grants from the University of Tasmania Graduate Research Scholarship and a Postgraduate Top-up scholarship from the Tasmanian Institute of Agriculture. We also acknowledge the collaborative research support of CopRice Feeds Pty Limited, Cobden, Victoria, for manufacturing the experimental oilenriched pellets to specification, field supervisory role of Mark Freeman, and Peter Nish of TasHerd Pty Limited, Hadspen, Tasmania for milk composition analysis.