
Immunotherapy: Open Access
Open Access
ISSN: 2471-9552

ISSN: 2471-9552
Research Article - (2022)Volume 8, Issue 4
Glioblastoma (GBM) is the most common and aggressive primary tumor of the central nervous system with high recurrence and extremely poor prognosis. Multiple recent studies have indicated a pivotal correlation between GBM prognosis and immune-related risk signature. Nevertheless, the potential value of Endothelial Cells (EC’s) Immune-Related Genes (EIRG’s) in prognosis, immune infiltration, and their correlation with therapeutic response to immunotherapy and TMZ chemotherapy remain obscure, especially in GBM. Here, we screened out 11 EIRG’s after intersecting the identified 59 GBM EC’s related prognostic genes and the identified 438 immune-related prognostic genes. A prognostic-related 6-EIRG’s signature was established through univariate Cox analysis and Least Absolute Shrinkage and Selection Operator (LASSO) Cox regression analysis, and patients in the high-risk group were significantly worse Overall Survival (OS) compared to those in the low-risk group. Additionally, univariate and multivariate Cox regression analysis confirmed that risk score was an independent predictor of OS in patients with GBM. The nomogram which comprised age, gender, IDH mutation status, radiation therapy, and risk score yielded a strong predictive ability of 0.5, 1, and 2 years OS for GBM patients. Our results demonstrate that the EIRG’s signature, which is associated with immune cell infiltration, may play a regulatory role in the immunobiological process of TIME (Tumor Immune Micro Environment). Prognostic-related 6-EIRG’s signature is a promising classification index for predicting the drug sensitivity to immunotherapy and TMZ chemotherapy, suggesting that EIRGs signature may serve as a biomarker to stratify patients who will benefit from immunotherapy and chemotherapy.
Glioblastoma; Endothelial cells; Immune infiltration; Prognosis signature
BBB: Blood-Brain Barrier; CGGA: Chinese Glioma Genome Atlas; ECs: Endothelial Cells; EIRGs: Endothelial Cells Immune-Related Genes; GBM: Glioblastoma; GO: Gene Ontology; GSEA: Gene Set Enrichment Analysis; GSVA: Gene Set Variation Analysis; IC50: Half-Maximal Inhibitory Concentration; IRGPs: Immune-Related Gene Pairs; KEGG: Kyoto Encyclopedia of Genes and Genomes; LASSO: Least Absolute Shrinkage and Selection Operator; MSI: Microsatellite Instability; OS: Overall Survival; ssGSEA: Single Sample Gene Set Enrichment Analysis; scRNA-seq: Single-cell RNA Sequencing; TCGA: The Cancer Genome Atlas; TIME: Tumor Immune Microenvironment; TMZ: Temozolomide; TIDE: Tumor Immune Dysfunction and Exclusion.
Glioblastoma (GBM) is the most common and aggressive primary tumor of the central nervous system, which is characterized by a high degree of intra-tumoral heterogeneity and resistance to treatment [1-3]. Despite recent advances in multimodal standard therapy, including surgical resection, radiotherapy, and Temozolomide (TMZ) chemotherapy, and immunotherapy, the prognosis of patients with GBM is still poor, with a median survival time of around 15 months [4-6]. So far, the alkylating drug Temozolomide (TMZ) is the mainstay of first-line chemotherapy for the treatment of GBM, but drug resistance to TMZ chemotherapy and drug delivery across the Blood-Brain Barrier (BBB) are the major obstacles to the effective treatment of GBM [7,8]. Additionally, growing evidence has indicated that immunotherapy has revolutionized the treatment of many solid-tumor malignancies and generated great hope for GBM, but without success until now [9-11]. Therefore, novel biomarkers that can predict the drug sensitivity to immunotherapy and TMZ chemotherapy in GBM should be sought. Meanwhile, the Tumor Immune Micro Environment (TIME) also plays a critical role in tumorigenesis, progression, therapeutic efficacy, and clinical outcomes of GBM patients [12-14]. Thus, elucidating the role of the components and signatures of the tumor immune microenvironment and the interaction between cancer cells and the TIME may help to understand the pathogenesis of tumors and guide the development of novel therapeutic interventions.
Cerebral Endothelial Cells (EC’s) are the major components of the BBB. EC’s are connected by complex tight junctions and exhibit very low levels of vesicular transcytosis, leading to a very low vascular permeability [15,16]. Additionally, EC’s exert the barrier characteristics of the BBB in binding and efflux of various small lipophilic foreign compounds and drugs through the specific transport network of efflux transporters, thus limiting the development and absorption of neurotherapeutic drugs [17]. Tumor heterogeneity complicates the treatment of patients with cancer [18], therefore, we should further understand how EC’s affect the prognosis of cancer and how it fosters the emergence of drug resistance, so as to provide novel breakthroughs and therapeutic strategies for targeted cancer therapy in the future. Of note, Xie et al. have explored the heterogeneity of the human BBB and its molecular alteration in GBM by scRNA-seq–based molecular atlas of human brain EC’s [19]. A previous study has revealed that the 1p/19 codeletion-associated immune prognostic signature could accurately predict the prognosis of glioma [20]. Moreover, our earlier study had demonstrated that Immune-Related Gene Pairs (IRGP’s) signature, which is associated with immune cell infiltration, can be used for individual OS predictions for glioma patients [21]. Nevertheless, little is known about how ECs-Immune-Related Genes (EIRG’s) affect GBM prognosis and how the relationship between EIRG’s and infiltrating immune cells in the TIME, and their correlation with therapeutic response to immunotherapy and TMZ chemotherapy.
This study was conducted with an aim to identify EIRG’s, whose expression is correlated closely with the prognosis of GBM patients in The Cancer Genome Atlas (TCGA) database. To further assess the prognostic value of these prognostic-related EIRG’s, univariate Cox regression analysis and LASSO Cox regression analysis were performed to construct a ECs-immune-related gene prognostic model to facilitate the prediction of GBM prognosis. Subsequently, a validation model from the Chinese Glioma Genome Atlas (CGGA) database was constructed to evaluate the feasibility of the risk signature based on the ECs-immune-related genes. Moreover, we analyzed the abundances of the tumor-infiltrating immune cells in GBM samples between the risk groups to explore the association between prognostic-related EIRG’s and the Tumor Immune Micro Environment (TIME). Finally, we explored these EIRG’s signature association with therapeutic response to immunotherapy and TMZ chemotherapy.
Data acquisition
The RNA-seq transcriptome data and corresponding clinical data of GBM patients were downloaded from The Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov/) and the Chinese Glioma Genome Atlas (CGGA, http://www.cgga.org.cn) databases. The 374 GBM endothelial cells (ECs) related genes were acquired from the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) database (GSE162631) and the 2333 immune-related genes were downloaded from the Immunology Database and Analysis Portal (ImmPort) database (https://www.immport.org/home).
Construction of EC-immune-related genes (EIRGs) risk model
Univariate Cox regression analyses with P-value<0.05 were performed to identify 59 GBM ECs related prognostic genes and 438 immune-related prognostic genes. To screen the EIRGs for GBM patients, the above-identified EC’s related prognostic genes and immune-related prognostic genes were intersected in the TCGA and CGGA datasets. From this, we identified a total of 11 prognostic EIRG’s, with a statistically significant correlation (P<0.05), which we developed as a potential risk signature for the selection of candidate risk EIRGs with the LASSO Cox regression analysis. The TCGA set was utilized to construct an EIRGs-related prognostic model, and the CGGA set was used to validate the prognostic accuracy of this established model. Finally, 6 EIRGs with their coefficients (β) were determined according to the minimum criteria. Risk score for each patient in the TCGA and CGGA cohorts were calculated according to the following formula: Risk score=(expression of gene A1*β1)+(expression of gene A2*β2)+(expression of gene A3*β3)+... (Expression of gene An*βn). Then, we divided patients into two groups: the high-risk group and the low-risk group based on the median value of the risk score. The Kaplan-Meier survival analysis was used to evaluate the differences in Overall Survival (OS) between high and low-risk score groups by using the "survival" R package.
Independence of the prognostic-related EIRGs risk model and nomogram analysis
Additionally, univariate and multivariate Cox regression analyses were used to determine whether the EIRGs-related risk scores were independent of other clinicopathological and molecular features (age, gender, IDH mutation status, radiation therapy, and risk score) as independent predictors for patients with GBM. Nomogram analysis for predicting the probability of 0.5-, 1-, and 2-year OS for GBM patients by integrating risk score and clinical features (age, gender, IDH mutation status, and radiation therapy) was constructed and verified in the TCGA cohort as well as CGGA cohort using the “rms” R package. Moreover, we plotted the calibration curve to validate the predictive accuracy and reliability of the nomogram. Afterward, heatmaps were drawn to investigate the correlation between EIRGs-related risk scores and clinicopathological and molecular features using the “Complex Heatmap” R package [22].
Evaluation of tumor microenvironment infiltration patterns and immune cells infiltration
The stromal score, immune score, estimate score, and tumor purity were calculated with the ESTIMATE algorithm using the “estimate” R package [23]. Subsequently, to investigate the immune infiltration landscape of GBM, we utilized the ssGSEA (Single Sample Gene Set Enrichment Analysis) to assess 23 types of tumor-infiltrating immune cells and further revealed a correlation between tumor-infiltrating immune cells and different risk groups. Subsequently, the 'GSVA' R package (v.1.38.2) was used for ssGSEA to evaluate immune-related functions or pathways between high-risk and low-risk groups [24]. And then, Gene Set Enrichment Analysis (GSEA) of prognosis-related EIRGs signature was performed for the assessment of the enrichment of biological processes and pathways among high-risk and low-risk groups [25]. The KEGG gene sets (c2.cp.kegg.v7.4.symbols.gmt), Go-Biological Processes (GO.BP) gene sets (c5.go.bp.v7.4.symbols.gmt), and HALLMARK gene sets (h.all.v7.4.symbols.gmt) were downloaded from the Molecular Signatures Database (MSigDB, http://www.gsea-msigdb.org/gsea/downloads.jsp) to run the GSEA analysis. The“clusterProfiler” R package (V3.18.1) was used to perform the top 10 functional annotations and pathway analysis on KEGG, GO, and hallmark pathway on EIRGs with an adjusted p<0.05, and FDR<0.05 [26].
Therapeutic response prediction
The Tumor Immune Dysfunction and Exclusion (TIDE) score of patients with GBM from the TCGA dataset was downloaded from the TIDE website (http://tide.dfci.harvard.edu/) for predicting the response to immunotherapy. Higher TIDE predictive scores were associated not only with greater potential of tumor immune evasion but also with worse immunotherapy response. The correlation between the identified EIRGs signature and Drug response was predicted by CellMiner (https://discover.nci.nih.gov/cellminer/home.do). To examine the sensitivity of each sample to chemotherapeutic response to TMZ, the "pRRophetic" R package was utilized to predict the half-maximal inhibitory concentration (IC50) of TMZ for the comparison of the different risk groups [27]. P<0.05 was considered statistically significant.
Identification of GBM endothelial cells (EC’s) immune-related genes
The detailed workflow of the risk model construction and downstream analysis is shown in Figure 1. As shown in the Venn diagram, a total of 11 prognostic EC’s immune-related genes (EIRG’s), which were indicated to have significant prognostic value (P<0.05), were screened out after intersecting the identified 59 GBM EC’s related prognostic genes and the identified 438 immune-related prognostic genes (Figures 1c and 2a). Subsequently, we performed to investigate the correlation among the identified EIRG’s with a threshold of correlation coefficient was 0.2. The correlation analysis results in this study showed that both SBDS and RBP1 were negatively correlated with PLXND1, while other EIRGs exhibited different positive correlation (Figure 2b).
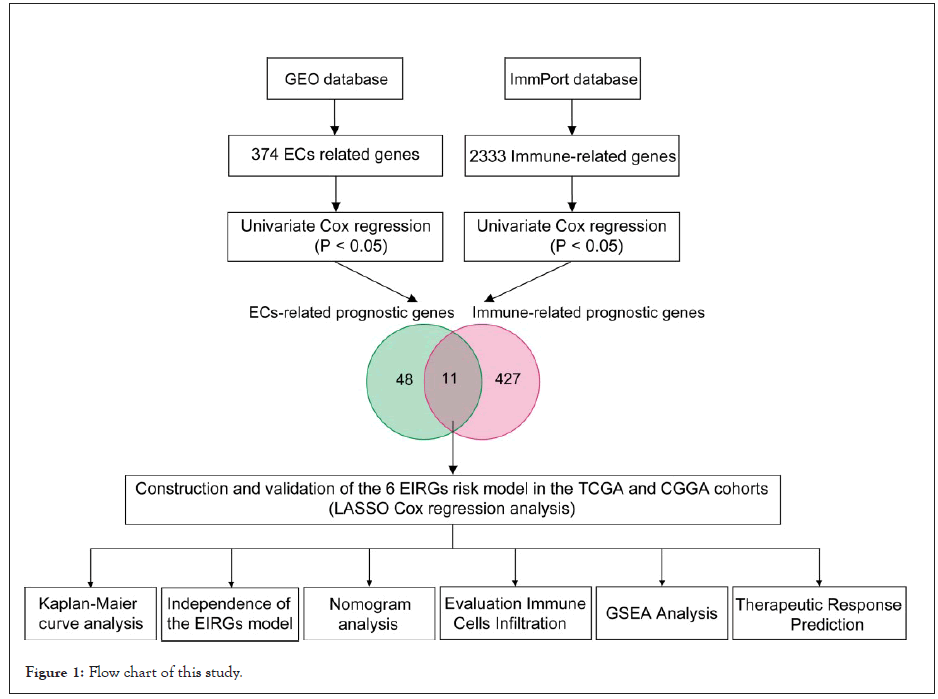
Figure 1: Flow chart of this study.
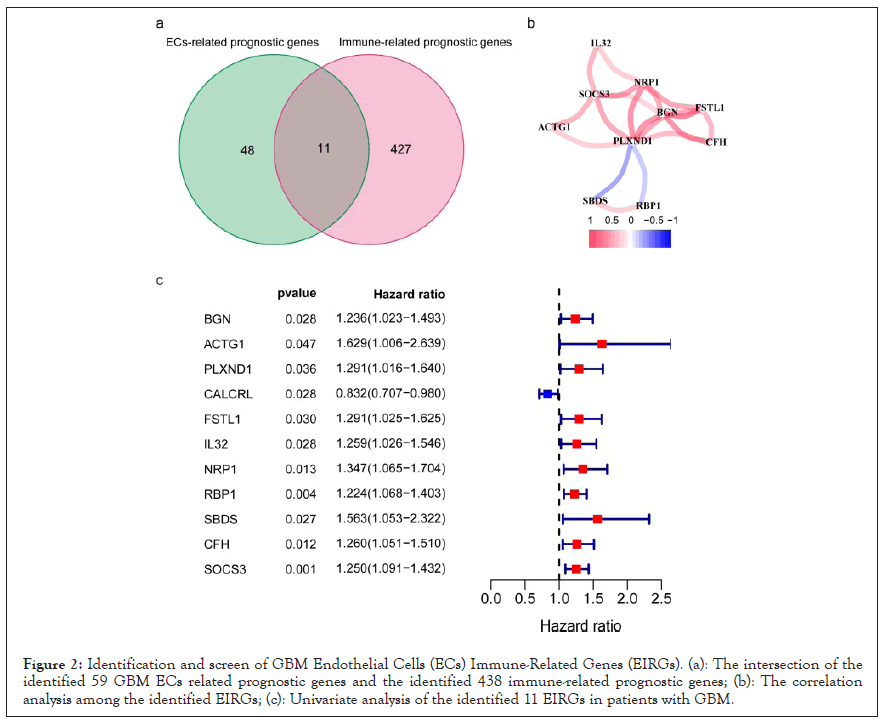
Figure 2: Identification and screen of GBM Endothelial Cells (ECs) Immune-Related Genes (EIRGs). (a): The intersection of the identified 59 GBM ECs related prognostic genes and the identified 438 immune-related prognostic genes; (b): The correlation analysis among the identified EIRGs; (c): Univariate analysis of the identified 11 EIRGs in patients with GBM.
Construction and validation of the prognostic-related 6-EIRG signature
To build the EIRG’s signature for forecasting the OS of GBM patients, we performed the least absolute shrinkage and selection operator (LASSO) Cox regression analysis on the basis of the 11 prognostic EIRGs in the TCGA cohort. Next, we identified 6 prognostic-related EIRGs (PLXND1, CALCRL, RBP1, SBDS, CFH, and SOCS3) to build the risk model, and the coefficients of these genes were used to calculate the risk score (Figure 3a). The risk score=PLXND1 × 0.0521+CALCRL × (-0.0292)+RBP1 × 0.0878+SBDS × 0.0912+CFH × 0.0656+SOCS3 × 0.0831. According to the median value of the risk scores, GBM patients were divided into the high-risk group and low-risk group, and then performed Kaplan-Meier survival analysis. Risk score, survival status distributions, and the relative expression standards of the 6 prognostic-related EIRG’s for each patient are shown in Figure 3b. The Kaplan-Meier survival analysis showed that patients in the high-risk group displayed worse Overall Survival (OS) compared to those in the low-risk group (P<0.001), indicating that the risk score could predict the prognosis in the TCGA cohort (Figure 3c). To validate the prognostic ability of the prognostic risk model, we calculated risk scores for GBM patients in the CGGA cohort using the same formula (Figure 3d). The results of the Kaplan-Meier survival curve in the CGGA cohort were consistent with our findings in the TCGA cohort: GBM patients in the high-risk group had significantly worse OS than those in the low-risk group (P<0.001), which made our results convincing (Figure 3e).
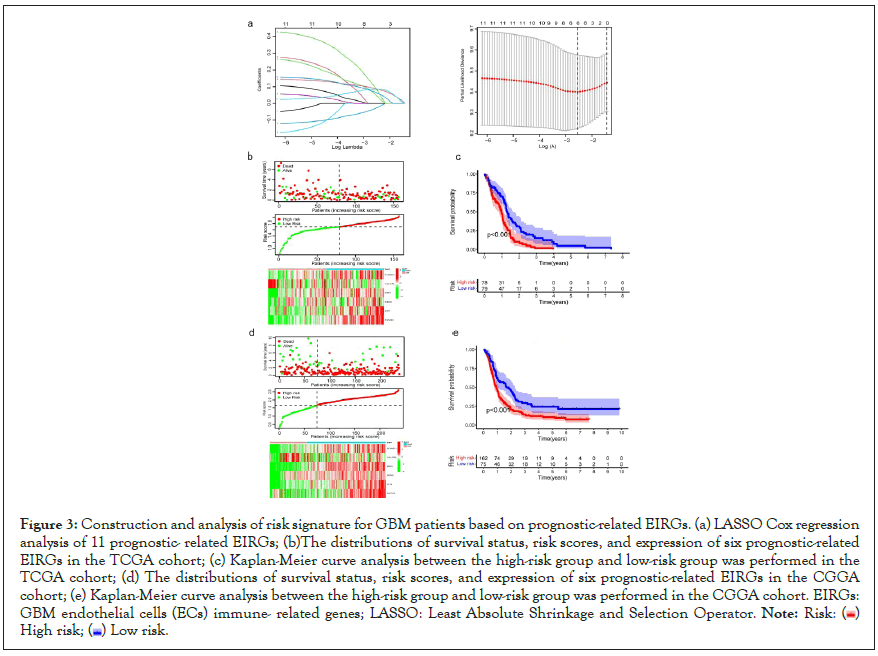
Figure 3: Construction and analysis of risk signature for GBM patients based on prognostic-related EIRGs. (a) LASSO Cox regression
analysis of 11 prognostic- related EIRGs; (b)The distributions of survival status, risk scores, and expression of six prognostic-related
EIRGs in the TCGA cohort; (c) Kaplan-Meier curve analysis between the high-risk group and low-risk group was performed in the
TCGA cohort; (d) The distributions of survival status, risk scores, and expression of six prognostic-related EIRGs in the CGGA
cohort; (e) Kaplan-Meier curve analysis between the high-risk group and low-risk group was performed in the CGGA cohort. EIRGs:
GBM endothelial cells (ECs) immune- related genes; LASSO: Least Absolute Shrinkage and Selection Operator.

Evaluation the correlation between risk score and the clinicopathological characteristics
The heat map shows clinical characteristics and the expression of the 6 prognostic-related EIRG’s in high and low-risk patients in the TCGA cohort (Figure 4a). We observed significant differences between the high and low-risk groups associated with IDH mutation status, stromal score, immune score, estimate score as well as tumor purity. To determine whether the prognostic value of the 6 prognostic-related EIRG’s signature-based risk score is independent of other clinical features, univariate and multivariate Cox regression analyses were performed to analyze with risk score and other clinical features, such as including age, gender, IDH mutation status, and radiation therapy. Univariate and multivariate Cox analysis indicated risk score remained to be an independent prognostic factor P<0.05 (Figures 4b and 4c).
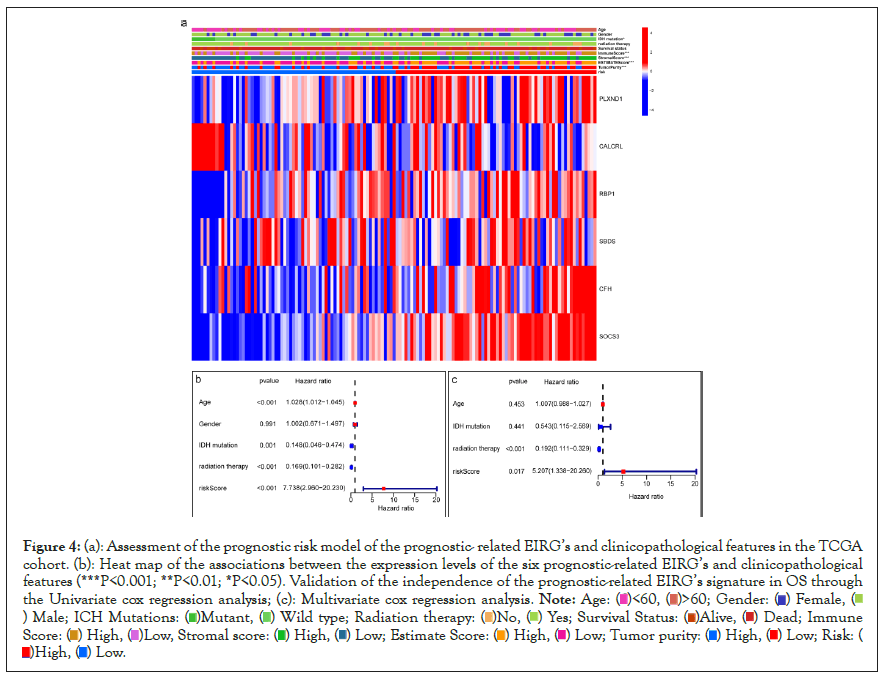
Figure 4: (a): Assessment of the prognostic risk model of the prognostic- related EIRG’s and clinicopathological features in the TCGA
cohort. (b): Heat map of the associations between the expression levels of the six prognostic-related EIRG’s and clinicopathological
features (***P<0.001; **P<0.01; *P<0.05). Validation of the independence of the prognostic-related EIRG’s signature in OS through
the Univariate cox regression analysis; (c): Multivariate cox regression analysis.
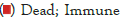
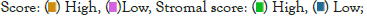
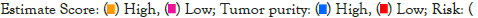
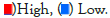
Nomogram establishment and evaluation
As shown in Figures 5a-5d, a nomogram for predicting 0.5, 1 and 2-year survival rates of GBM patients was established according to age, gender, IDH mutation status, radiation therapy, and risk score. The C-index of the prognostic nomogram was 0.707, and the calibration curve of the 0.5, 1 and 2-year OS indicated that our nomogram model had a good predictive ability. Importantly, we found similar results in the CGGA cohort (Figures 5e-5g).
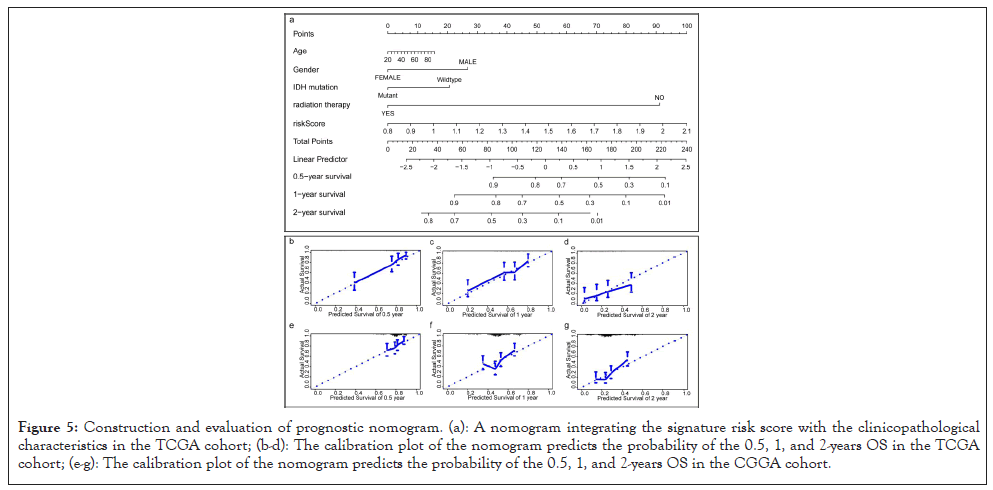
Figure 5: Construction and evaluation of prognostic nomogram. (a): A nomogram integrating the signature risk score with the clinicopathological characteristics in the TCGA cohort; (b-d): The calibration plot of the nomogram predicts the probability of the 0.5, 1, and 2-years OS in the TCGA cohort; (e-g): The calibration plot of the nomogram predicts the probability of the 0.5, 1, and 2-years OS in the CGGA cohort.
Estimation of the tumor immune microenvironment using the prognostic-related EIRGs risk model
As shown in Figure 6a, the immune, stroma, and ESTIMATE scores were significantly higher (p<0.05) whereas tumor purity was lower in the high-risk group compared with the low-risk group (p<0.05). To elucidate the difference in the immune microenvironment of GBM, we analyzed immune cell infiltrating density between high and low-risk groups. The results indicated that high immune infiltration was strongly related to the high-risk group (Figure 6b). Consistent with our previous results, the risk score of the prognostic model had a significant positive correlation with immune cells infiltration, including regulatory T cell (Treg), Th1 (T helper cell, type 1), activated dendritic cell, macrophages, Mast cell, NK natural killer T cells (NKT), MDSCs, and neutrophils (Figure 6c). Notably, the results from the correlation analyses showed that the risk score was positively correlated with activated CD4+ T cells, activated CD8+ T cells, activated B cells, activated dendritic cells, macrophages, regulatory T cells (Treg), and natural killer cells (P<0.05) (Supplementary Figure 1). Among the 29 immune gene sets, immune-related functional cells such as activated dendritic cells (aDCs), B cells, DCs, Interdigitating Dendritic Cells (IDCs), macrophages, neutrophils, T helper cell, follicular helper T cells (Tfh), Th1 cells, Th2 cells, and regulatory T cells (Treg) showed higher ssGSEA scores in the high-risk patients (Figure 6d). Similarly, immune pathways such as Antigen-Presenting Cell (APC) co-inhibition, APC co-stimulation, CC Chemokine Receptor (CCR), Check-point, cytolytic activity, Human Leukocyte Antigen (HLA), inflammation-promoting, para-inflammation, T-cell co-inhibition, T-cell co-stimulation, Type II IFN Response, and Tumor-Infiltrating Lymphocytes (TIL) exhibited higher ssGSEA scores in the high-risk group (Figure 6d). Additionally, similar results were obtained in the CGGA cohort of GBM patients (Supplementary Figures 2 and 3). These data indicated that the 6 prognostic-related EIRG’s signature were highly associated with immune infiltration.
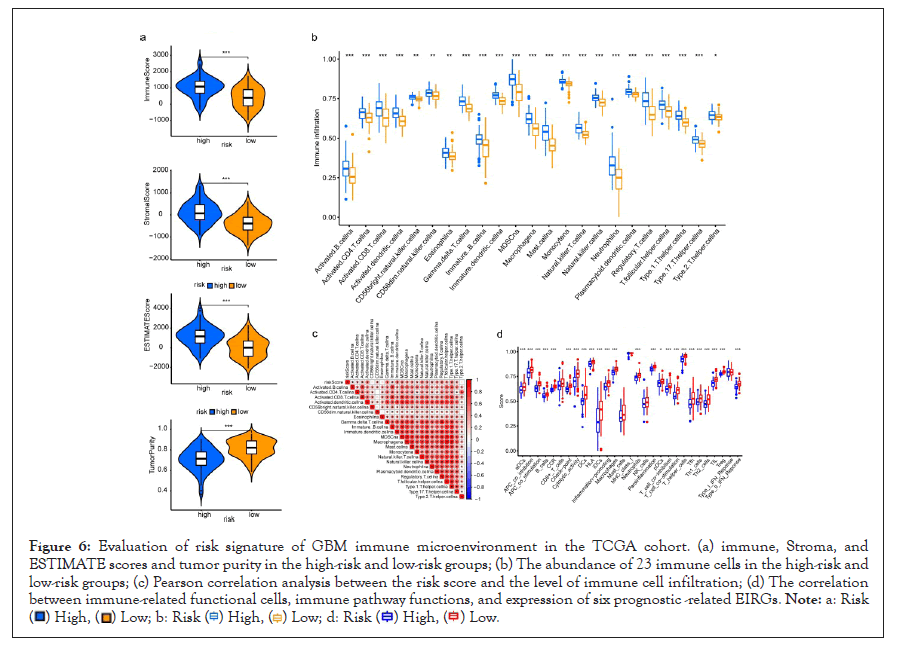
Figure 6: Evaluation of risk signature of GBM immune microenvironment in the TCGA cohort. (a) immune, Stroma, and
ESTIMATE scores and tumor purity in the high-risk and low-risk groups; (b) The abundance of 23 immune cells in the high-risk and
low-risk groups; (c) Pearson correlation analysis between the risk score and the level of immune cell infiltration; (d) The correlation
between immune-related functional cells, immune pathway functions, and expression of six prognostic -related EIRGs.
GSEA analysis of 6-EIRG’s signature
Next, GSEA analysis was performed to assess the involvement of biological processes, immune-related pathways, and cancer-related hallmarks of each GBM sample. The top 10 KEGG pathways, GO biological processes (GOBP), and hallmark gene sets were identified based on the P-value<0.05 in the TCGA and CGGA cohorts. The GSEA analysis showed that most of the KEGG pathways, GOBP, and hallmark gene sets enriched in the high-risk group were associated with the immune and inflammatory responses as well as the cancer-related pathway, including Toll-like receptor signaling pathway, JAK/STAT signaling pathway, NOD-like receptor signaling pathway, IL-2-STAT5 signaling pathway, IL-6-JAK-STAT3 signaling pathway, TNFA signaling via NFKB, inflammatory response, activation of the immune response, and others in the TCGA and CGGA cohorts (Figures 7a and 7b). These findings indicate that prognostic-related EIRG’s may take part in regulating the tumor immune environment and immunologic biological processes of GBM.
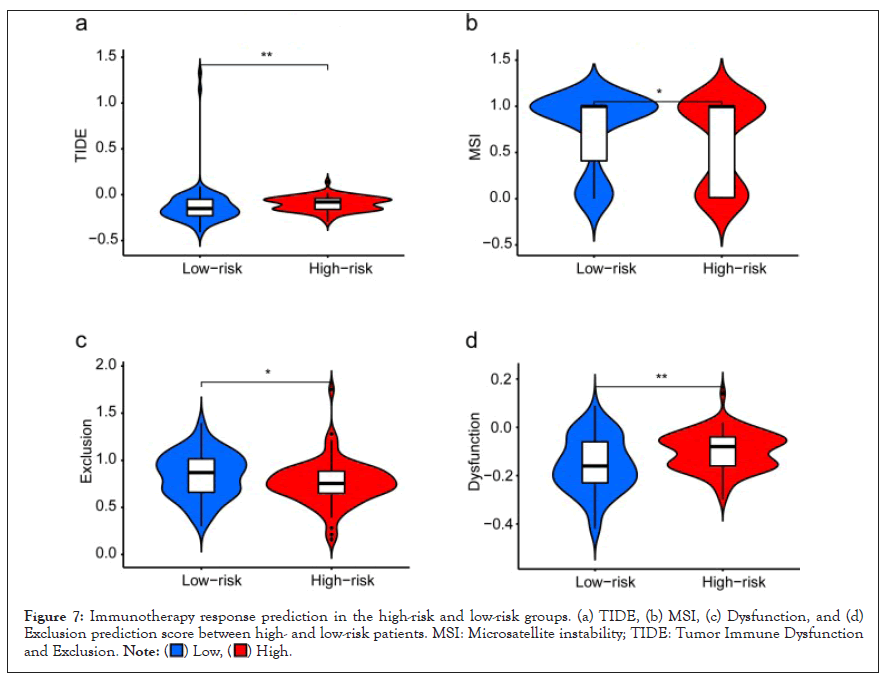
Figure 7: Immunotherapy response prediction in the high-risk and low-risk groups. (a) TIDE, (b) MSI, (c) Dysfunction, and (d)
Exclusion prediction score between high- and low-risk patients. MSI: Microsatellite instability; TIDE: Tumor Immune Dysfunction
and Exclusion.
Sensitivity of different therapies in the high and low-risk groups
Next, we further predicted the likelihood of response to immunotherapy in the high-risk and low-risk groups using the TIDE algorithm. TIDE scores were significantly higher in the high-risk group compared with the low-risk group, which indicated the low-risk group was much more sensitive to immunotherapy (Figure 8a). Subsequently, we found that GBM patients with high-risk group exhibited higher Microsatellite instability (MSI) and Dysfunction, and lower expression of Exclusion compared to the patients with the low-risk group (Figure 8b-8d). Additionally, a significant correlation relationship between the expression level of the key EIRGs and chemotherapy drug sensitivity are displayed in Figure 8 and ranked by the p-value, selected by p<0.05. Notably, SOCS3 was positively correlated with the sensitivity of Bleomycin, Sonidegib, Irofulven, and Olaparib and the resistance of Selumetinib, Encorafenib, Cobimetinib, and ARRY-162. Highly expressed PLXND1 were more resistant to Palbociclib (Cor=-0.366, p=0.004) and METHOTREXATE (Cor=-0.355, p=0.005) (Figure 9).
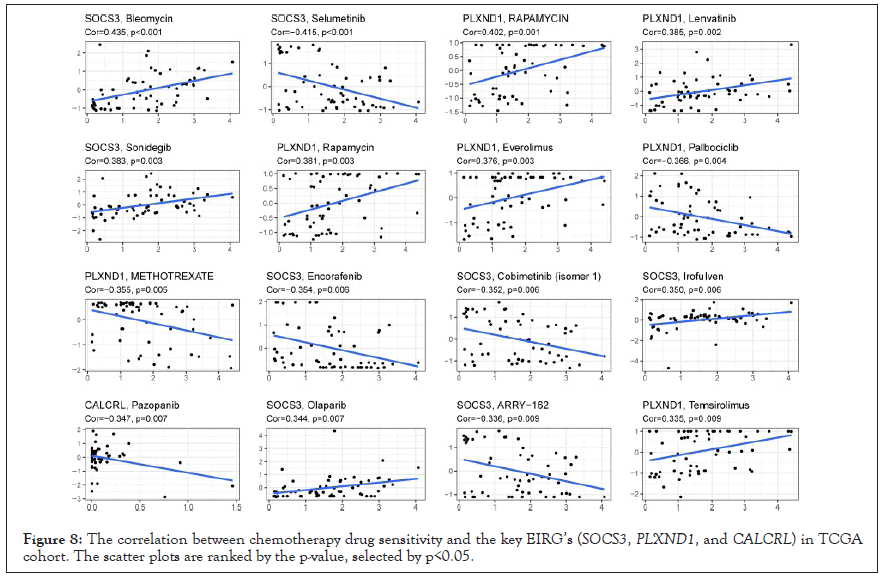
Figure 8: The correlation between chemotherapy drug sensitivity and the key EIRG’s (SOCS3, PLXND1, and CALCRL) in TCGA cohort. The scatter plots are ranked by the p-value, selected by p<0.05.
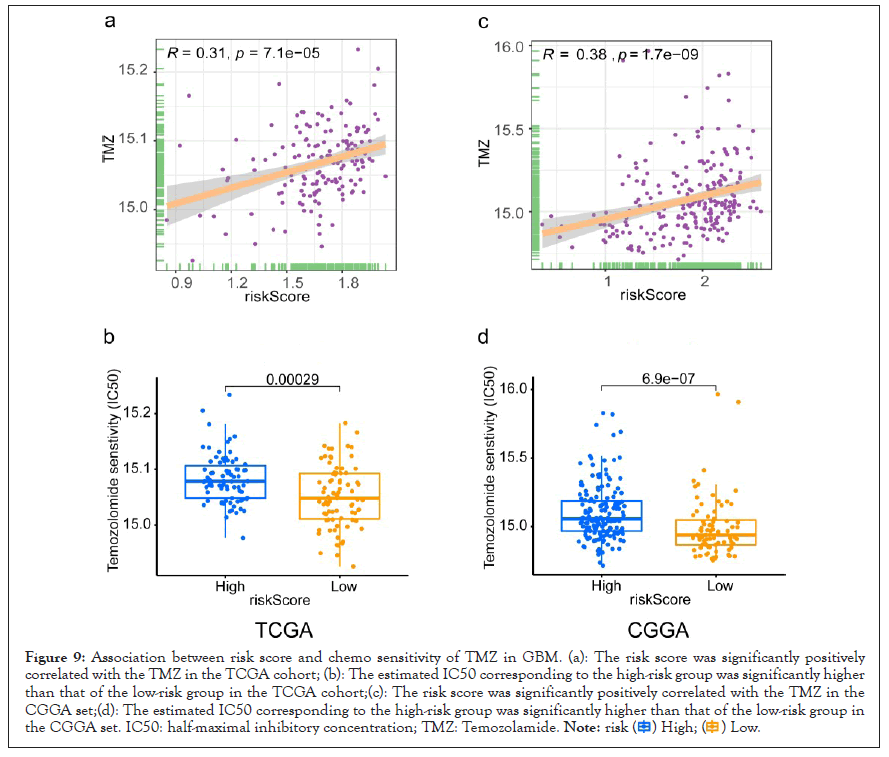
Figure 9: Association between risk score and chemo sensitivity of TMZ in GBM. (a): The risk score was significantly positively
correlated with the TMZ in the TCGA cohort; (b): The estimated IC50 corresponding to the high-risk group was significantly higher
than that of the low-risk group in the TCGA cohort;(c): The risk score was significantly positively correlated with the TMZ in the
CGGA set;(d): The estimated IC50 corresponding to the high-risk group was significantly higher than that of the low-risk group in
the CGGA set. IC50: half-maximal inhibitory concentration; TMZ: Temozolamide.
Afterwards, we further explore the differences in the treatment response to temozolomide (TMZ) chemotherapy in the high and low-risk groups. The risk score was significantly positively correlated with the TMZ (P<0.05). The estimated IC50 corresponding to the low-risk group was significantly lower than that of the high-risk group, indicating that GBM patients from the low-risk group were more sensitive to TMZ P<0.05. Notably, we observed similar results in the CGGA cohort. These results indicated that the prognostic-related EIRG’s signature could predict the treatment response to immunotherapy and TMZ chemotherapy, and guide effective therapy.
GBM is the most aggressive and lethal primary brain tumor with heterogeneous clinical features and a generally poor clinical outcome [28]. The treatment of GBM remains a great clinical challenge using temozolomide (TMZ) based on conventional chemotherapy and is mainly due to drug resistance [29]. Previous studies have reported that Xie et. al have performed scRNA-seq on GBM EC’s to characterize the heterogeneity of gene expression signatures of different ECs, which will provide key information for designing rational therapeutic regimens and optimizing drug delivery [19]. However, little is known about the prognostic-related EIRGs signature with tumor immune microenvironment and drug sensitivity to immunotherapy and TMZ chemotherapy. In this study, we successfully constructed a prognostic risk signature with six identified prognostic-related EIRG’s (PLXND1, CALCRL, RBP1, SBDS, CFH, and SOCS3), which divided GBM patients into high and low-risk groups. Multivariate Cox regression analysis showed that prognostic-related EIRGs signature-based risk score was an independent prognostic factor for OS (HR: 5.207; 95% CI: 1.338-20.260; P<0.05) after adjusting for other clinical variables. Moreover, combining risk score with age, gender, IDH mutation status, and radiation therapy, we constructed a nomogram, and it had a robust ability to predict 0.5-year, 1-year, and 2-year OS in GBM patients.
Tumor formation involves the co-evolution of neoplastic cells together with the TME surrounded by extracellular matrix, tumor vasculature, and immune cells, which may constitute the basis of tumor micro environmental heterogeneity due to the diversity of these cells [30]. Numerous studies had confirmed that immune infiltration is related to the prognosis of GBM, and tumor immune microenvironment significantly influences therapeutic response and clinical outcome [31,32]. Intratumor heterogeneity remains a major obstacle for more effective anti-cancer therapy and personalized medicine [33]. Multiple recent studies have shown that the heterogeneity of stromal cells and immune cells in the TIME also plays a critical role in cancer cell proliferation, angiogenesis, immune evasion, metastasis, and responses to treatment [34]. In our study, we have demonstrated that patients in the high-risk group had higher immune, stroma, and ESTIMATE scores, lower tumor purity, and a higher abundance of immune cell infiltration. Additionally, the GSEA analysis for the 6 EIRG-signature suggested that patients in the high-risk group were significantly enriched in biological processes, immune-related pathways, and cancer-related hallmarks, including immune responses (Toll-like receptor signaling pathway, TNFA signaling via NFKB, IL2-STAT5 signaling, IL6-JAK-STAT3 signaling, inflammatory response, and interferon-gamma response), cellular stress (hypoxia and apoptosis), and signal transduction (JAK/STAT signaling pathway and cytokine-cytokine receptor interaction). Notably, JAK/STAT signaling pathway plays an important role in immune regulation, apoptosis, drug resistance, tumor proliferation, migration, and invasion in glioma [35,36]. Moreover, previous studies also demonstrated that hypoxia may represent an early sign for GBM recurrence, which might become useful in the development of new combined diagnostic-therapeutic approaches of recurrent GBM [37]. These results indicated that the prognostic-related EIRG’s risk signature was closely associated with immunologic biological processes and the regulation of tumor immune microenvironment in GBM.
Therapeutic resistance reflects active tumor evolution, and environment-mediated resistance reflects the dynamic interaction between tumor cells and their surrounding microenvironment [38]. A growing body of evidence has demonstrated that Temozolomide (TMZ) is the most commonly used chemotherapeutic drug used to treat GBM, and its function is mainly achieved by inducing DNA damage in tumor cell [39,40]. Nevertheless, most GBM patients are resistant to the drug TMZ [41]. In our study, we found that the estimated IC50 corresponding to the high-risk group was significantly higher than that of the low-risk group, suggesting that GBM patients from the high-risk group based on prognostic-related EIRG’s could be more resistant to TMZ chemotherapy drug, which may partially explain worse survival outcomes of patients in the high-risk group. Interestingly, patients in the low-risk group who exhibited lower TIDE scores showed more likely effectiveness for immunotherapy than the high-risk group, which indicated that patients from the high-risk group might have a higher chance of anti-tumor-immune escape. Our analysis demonstrates that prognostic-related 6-EIRGs signature is a strong discriminative tool for examining the sensitivity of GBM patients to immunotherapy and TMZ chemotherapy. Recent clinical and experimental studies have demonstrated that the neoadjuvant administration of PD-1 blockade might generate enhanced antitumor immune responses and induce functional activation of tumor-infiltrating lymphocytes, which may represent a promising strategy in the treatment of GBM [42,43]. Additionally, Park et al. have reported that PD-1 blockade induces a long-term therapeutic response, and combination with TMZ further enhances antitumor efficacy [44]. Thus, in our study, by exploring the potential relationship between the risk stratification and drug sensitivity to immunotherapy and TMZ chemotherapy, we can further provide new insights and achieve optimal clinical benefit for the combination of targeted immunotherapy and TMZ chemotherapy in the GBM.
Remarkably, we are also aware of some limitations in this study. First, our prognostic model was constructed based on open accessed databases without our cohort. Second, the biological and regulatory mechanism of prognostic-related EIRGs in the GBM immune microenvironment and their interaction with tumor-infiltrating immune cells needed to be further verified using more in vivo and in vitro experiments. Third, there were no recruited cohorts to verify the prognostic value of our prognostic-related EIRG’s risk signature and drug sensitivity to immunotherapy and TMZ chemotherapy in high‑ and low‑risk groups.
Collectively, our study provides a comprehensive insight for revealing the important role of prognostic-related EIRG’s in the prognosis, tumor immune microenvironment, and drug sensitivity to immunotherapy and TMZ chemotherapy of GBM, which provides a new research strategy for the early diagnosis, treatment, and prognosis of GBM. We constructed a prognostic-related 6-EIRG’s signature with good prognostic value and which can serve as an independent prognostic indicator for GBM. Notably, this is the first study to explore the relationship between prognostic-related EIRG’s and TIME, and drug sensitivity to immunotherapy and TMZ chemotherapy in the GBM. Our results indicate that the risk stratification based on risk signature might help distinguish GBM patients that could benefit from immunotherapy and chemotherapy drug TMZ.
Sponsored by Key Research and Development Program of Jiangsu Province (No.BE2019652), Changzhou international cooperation program (CZ20200039) and Development Program of Changzhou City (CE20205024).
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
Citation: Liu F, Xie Q, Zhang Y, Wang H, Huang W (2022) Endothelial Cells Immune-related Genes Risk Signature Correlated with Tumor Immune Microenvironment Predicts Therapeutic Response and Prognosis in Glioblastoma. Immunotherapy. 8:196.
Received: 04-Jul-2022, Manuscript No. IMT-22-17478; Editor assigned: 07-Jul-2022, Pre QC No. IMT-22-17478 (PQ); Reviewed: 22-Jul-2022, QC No. IMT-22-17478; Revised: 28-Jul-2022, Manuscript No. IMT-22-17478 (R); Published: 05-Aug-2022 , DOI: 10.35248/2471-9552.22.8.196
Copyright: © 2022 Liu F, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.