
Immunotherapy: Open Access
Open Access
ISSN: 2471-9552

ISSN: 2471-9552
Research Article - (2022)Volume 8, Issue 4
Background: Head and Neck Squamous Cell Carcinoma (HNSCC) is the most prevalent malignant tumor of the maxillofacial area. ARGs (Autophagy-Related Genes) are involved in the cell cycle and translation, which will fuel carcinogenesis.
Material and methods: TCGA data for ARGs in HNSCC were analyzed. The functional role of the ATG5 in HNSCC was investigated through ATG5 knockdown cell lines.
Results: According to the TCGA data, ATG5 was shown to be an adverse prognostic marker. Regardless of tumor stage, grade, or clinical characteristics, HNSCC patients with high ATG5 expression had a poorer survival rate. ATG5 knockdown suppressed the malignant features of HNSCC cell lines. The mechanism study demonstrated ATG5 knockdown could prevent the malignant phenotype of HNSCC by reversing the cell cycle and translation. One of the systems influencing cell cycle control in ATG5-dependent ways might include HDAC2, TTK, and CDK1. MRPL18, MRPL13, and MRPS14 might all be implicated in ATG5-dependent translation regulation.
Discussion and conclusion: The current research suggests that ATG5 is a key player in cell cycle and translation regulation, as well as poor prognosis in HNSCC.
Chemotherapy; Cell cycle; Translation; Malignancies
Head and Neck Squamous Cell Carcinoma (HNSCC) is the most prevalent malignant tumor of the maxillofacial area, accounting for more than 95% of head and neck malignancies. It is highly aggressive and has a high mortality rate, with poor overall survival rate of approximately 50% [1]. Surgery, radiotherapy, chemotherapy, or a combination of multiple modalities are all alternatives for treating head and neck squamous carcinoma, but there is no standard treatment strategy. Because the head and neck area converge with many important organs, such as the eyes, nose, and tongue, and patients' compliance with treatment is reduced, surgery may have negative effects on patients' appearance, diet, and other aspects, even though it can effectively inhibit the development of the disease [2]. The State Drug Administration has approved Cetuximab, an epidermal growth factor receptor inhibitor, and Pembrolizumab, an anti-PD-1/PD-L1 monoclonal antibody, for the first-line therapy of head and neck squamous carcinoma [3]. However, when administered alone, these medications are ineffective, and even when paired with radiotherapy or chemotherapy, they have minimal therapeutic effects. In the recent decade, the death rate of head and neck squamous carcinoma has not improved significantly. As a result, developing extremely sensitive tools for early diagnosis of HNSCC and identifying effective therapy targets for HNSCC are critical.
Autophagy, a type II programmed cell death, plays a crucial role in cancer along with Autophagy-associated (ATG) proteins. Many ATG protein molecules and their core complexes, such as the autophagy-specific class III PI3K complex, ULK1/2 kinase core complex, ATG12, ATG9A transporter system and LC3 ubiquitin-like coupling system, provide multiple activities of autophagic pathways implicated in autophagy induction, nucleation, elongation, growth, merging, and degradation, are still debated to this day [4,5]. Autophagy operates as a tumor suppressor or promoter in various situations and stages of cancer development. In the early stages of carcinogenesis, autophagy acts as a survival path and quality method of control, preventing tumorigenesis and inhibiting tumor growth. For example, boosting autophagic flux to decrease mammary carcinogenesis by interfering with the interaction of Beclin 1 and Bcl-2 has been found to support autophagy's tumor suppressive effect in the MMTV-Neu mice model [6]. In the Beclin1+/- mouse model, however, the spontaneous frequency of malignant tumors was greater, supporting autophagy as a tumor suppressor mechanism. Autophagy, on the other hand, functions as a dynamical degradation and cycling system that can help established tumors survive and develop by scavenging harmful oxygen free radicals and broken proteins, preserving mitochondrial function, supporting metabolic, as well as surviving under stress. Autophagy has been implicated in the survival of cancer cells in numerous studies. Autophagy has been linked to increased tumor aggressiveness through the promotion of metastases. Furthermore, autophagy may impair the therapeutic efficiency of most chemotherapeutic drugs as a cellular defense mechanism. While ATG proteins, the core proteins of mammalian autophagy, are important in the regulation of autophagy, no studies have been done on the role of particular ARGs in the development of head and neck squamous carcinoma.
In our study, we sought to evaluate the clinical predictive significance of basic ARG in HNSCC. In HNSCC, we discovered that a higher expression of ATG5 was found to be a significant poor prognostic marker. We also conducted additional research to confirm the role of ATG5 in HNSCC. HNSCC cell lines with ATG5 knockdown had their malignant characteristics suppressed [6]. ATG5 may be involved in regulating cell cycle and translational activities in HNSCC cells, according to further mechanistic analysis. Our data imply that ATG5 is a poor prognostic biomarker and could be applied to target HNSCC therapy [7].
Cell lines
HNSCC cell lines SCC1, SCC9, SCC15, and SCC25 were acquired from Type Culture Collection of the Chinese Academy of Sciences in Shanghai, China. At a 37°C incubator with 5% CO2, all the cells were grown in Dulbecco's Modified Eagle's Medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin [8]. ATG5 gene expression was inhibited by two sequences of shRNAs, pLVX-ATG5-shRNA1 and pLVX-ATG5-shRNA2 (PPL, PPL00042).
Western blotting assay
Centrifuged at 1,500 rpm, cells in culture were collected and washed twice with ice-cold PBS to eliminate leftover media. To extract protein, RIPA lysis solutions were added to cell pellets. Protein concentrations in lysates were determined using the Pierce BCA Protein Assay Kit, then SDS loading buffer (5X) was added and boiled at 95°C for 10 minutes. Protein samples were run on a 10% gradient gel SDS-PAGE and then transferred to a PVDF membrane. The membrane was blocked for 1 hour at room temperature in 5% milk before being treated with primary antibodies overnight at 4⸰C [9]. The membrane was washed three times with TBST the next day and incubated with secondary antibodies for one hour at room temperature. Enhanced Chemiluminescence (ECL) was used to image membranes and the following primary antibodies were used: ATG5, GAPDH, HDAC2 (Cell Signaling Technology, 2540S), TTK, CDK1 (Abcam, ab5467), MRPL18, MRPL13, MRPS14.
Flow cytometry analysis
Cells were planted in 6-well plates at a density of 0.5-1 × 106 cells per well. Centrifuged cells were collected, rinsed in cold PBS, and suspended in 1 ml of 75% ethanol in PBS and refrigerated overnight at -2°C. Cells were centrifuged again and washed twice with cold PBS [10]. After that, the cells were resuspended in PBS containing 20 mg/ml Propidium Iodide (PI), 0.1% Triton X-100 (Sigma, X100-100 ml), and 0.1 mg/ml RNase A and incubated at 37 for 45 minutes. After incubation, cells were kept at 4°C until flow cytometry analysis.
Cell migration assay
Briefly, cells were pre-treated in serum-free medium overnight before seeded onto the upper chamber of the insert. In the bottom chamber, media with 20% serum was employed. A cotton-tipped applicator was used to remove cells that remained on the top of the membrane after 24 hours. Cells that had migrated to the insert's bottom were fixed in 70% ethanol and stained with 0.1% Crystal Violet. Migrated cells were imaged with an inverted microscope, quantified with image [11].
Translation efficiency analysis
For translation efficiency analysis, cells were treated with puromycin at a final concentration of 1 M for 30 minutes at 37°C. The cells were then harvested and lysed in RIPA buffer with 1 mM PMSF and protease inhibitor. Anti-puromycin was used to detect the cell translation efficiency by Western blot.
Polysome profile assay
The polysome profiling test was carried out as previously described. In brief, cells were seeded in 15 cm plates (about 10 × 106 cells per plate). The cells were subsequently treated with 100 ug/ml actinomycin and incubated for 10 minutes at 37°C. The cells were then harvested, and cell extract was fed into a sucrose gradient ranging from 10% to 50% using a BioComp gradient station [12]. The gradient was revolved in the TH-641 rotor for 3 hours at 36000 rpm and 4°C. Finally, Bio Comp gradient station was used to assess the gradient (260 nm) and gathered the components.
TCGA data
UCSC XENA (http://xena.ucsc.edu/) was used to get RNA-seq readings and clinical data from TCGA patients with head and neck squamous cell carcinoma. The study eliminated samples that did not have clinical survival status follow-up data. These ARGs, which were discovered as significant autophagy regulators, were required for autophagosomes formation, and were analyzed further [13]. Throughout the project, the main tools for data analysis were R studio and R programming (4.03).
Survival analysis
The "surv cutpoint" function in the "survminer" package was used to find the best ARG cutoff values according to the expressions Transcript Per Million reads (TPM) value, survival time, and survival status. Patients having relative values above or below the optimal threshold for each gene expression level were separated into high or low groups. The difference in OS between the high and low groups was then investigated using Kaplan-Meier (KM) survival analysis with log-rank test.
Statistical analysis
Statistical significance was calculated using the Graph Pad Prism 8 software and R studio software, and p<0.05 was used to determine statistical significance [14]. Unless otherwise stated, data is provided as mean ± SD. Statistical comparisons were analyses by unpaired Student t-test with two-tailed p value. The KM method and the log-rank test were used to conduct the survival analysis.
Prognosis of ARGs in HNSCC
In HNSCC, high levels of ATG5 and ATG12 expression were linked to a poor prognosis, while patients with ATG2A, ATG3, ATG4B, ATG4C, ATG7, and ATG9B overexpression have a longer OS and a better 5-year survival rate. According the data, ATG5 and ATG12 has been discovered as a risk prognostic predictor, whereas other six ARGs have been identified as positive prognostic indicators. Furthermore, according to KM survival analysis, ATG5 expression can accurately predict the prognosis of patients with HNSCC with varied clinical phases, Tumor stage, and Tumor grade at diagnosis. Taken together, ATG5 seems to be the best candidate for further analysis.
ATG5 expression was up regulated and correlated with malignant phenotype in HNSCC
Firstly, ATG5 mRNA expression was analyzed between HNSCC tissues and normal tissues according to the TCGA database to see if ATG5 plays a role in HNSCC. ATG5 was shown to be up regulated in tumor tissues according to TCGA data (Figure 1A). ATG5 expression was also different depending on clinic pathologic factors: ATG5 was elevated with tumor stage 4 compared to lower tumor stage (Figure 1B). And ATG5 expression was also linked to tumor grade (Figure 1C). Besides, ATG5 expression was higher in patients with HPV infection than HPV negative, indicating that ATG5 serves as a risk factor in HNSCC (Figure 1D).
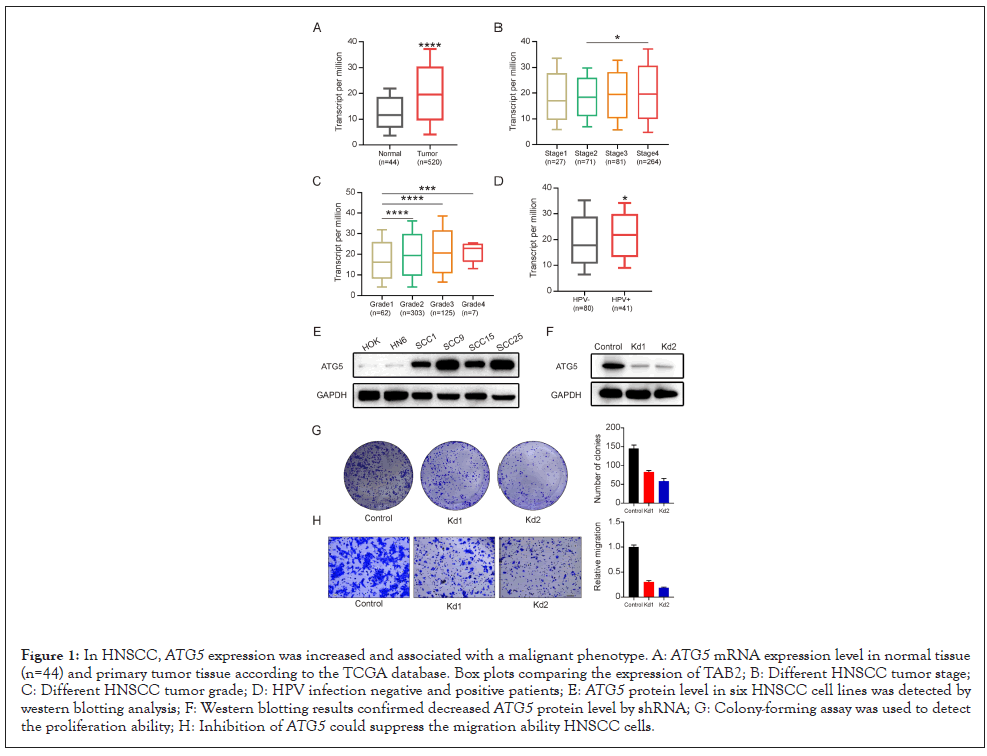
Figure 1: In HNSCC, ATG5 expression was increased and associated with a malignant phenotype. A: ATG5 mRNA expression level in normal tissue (n=44) and primary tumor tissue according to the TCGA database. Box plots comparing the expression of TAB2; B: Different HNSCC tumor stage; C: Different HNSCC tumor grade; D: HPV infection negative and positive patients; E: ATG5 protein level in six HNSCC cell lines was detected by western blotting analysis; F: Western blotting results confirmed decreased ATG5 protein level by shRNA; G: Colony-forming assay was used to detect the proliferation ability; H: Inhibition of ATG5 could suppress the migration ability HNSCC cells.
Then, six HNSCC cell lines were chosen and their HNSCC protein levels analyzed to study the function of ATG5 as a possible oncogene in HNSCC cells. One HNSCC cell line (SCC25) was found to have greater levels of expression (Figure 1E). Then we used two shRNAs aimed at ATG5 to treat SCC25, and the results revealed that protein levels were significantly reduced (Figure 1F). The clony formation experiment revealed that ATG5 downregulation significantly reduced cellular clony formation capacity (Figure 1G). Importantly, according to the results of the trans well migration experiment, silencing of ATG5 decreased HNSCC migratory potential (Figure 1H). Therefore, our results revealed that lowering ATG5 expression might stop HNSCC cells from proliferating and migrating [15].
Co-expression genes correlated with ATG5 in HNSCC
We investigated the ATG5 co-expressed genes in HNSCC patients based on the TCGA data, in which the statistical method was Pearson correlation coefficient. 4668 genes were identified to positively associate with ATG5, while 3724 genes were negatively associated. The top 50 genes that are positively or negatively correlated with ATG5 were shown on the heat map. Then the biological pathway of the top ATG5-related co-expressed genes was then investigated using KEGG enrichment analysis. The ribosome and the cell cycle are linked to ATG5 co-expressed genes. ATG5 expression was also shown to be positively associated with HDAC2, TTK, CDK1 (cell cycle pathway related genes) and MRPL13, MRPL18 MRPS13 (ribosome related genes).
ATG5 knockdown reversed cell cycle and translation of HNSCC cells
By cell cycle distribution assay, ATG5 deletion had a significant negative effect on the distribution of S phase of HNSCC cells when compared to the control group (Figure 2A). Subsequently, the puromycin and polysome profiling assay were used to determine the involvement of ATG5 in translation. Similarly, the results indicated that down regulation of ATG5 markedly reduced the whole protein synthesis in HNSCC cells (Figures 2B and 2C). Further investigation found that knocking down ATG5 in SCC25 cells down regulated the expression of cell cycle related markers (HDAC2, TTK, and CDK1), ribosome-related proteins (MRPL18, MRPL13, and MRPS14) (Figure 2D). In general, suppression of ATG5 could inhibit proliferation and migration in HNSCC cells by reversing the cell cycle and translation process through specific pathways.
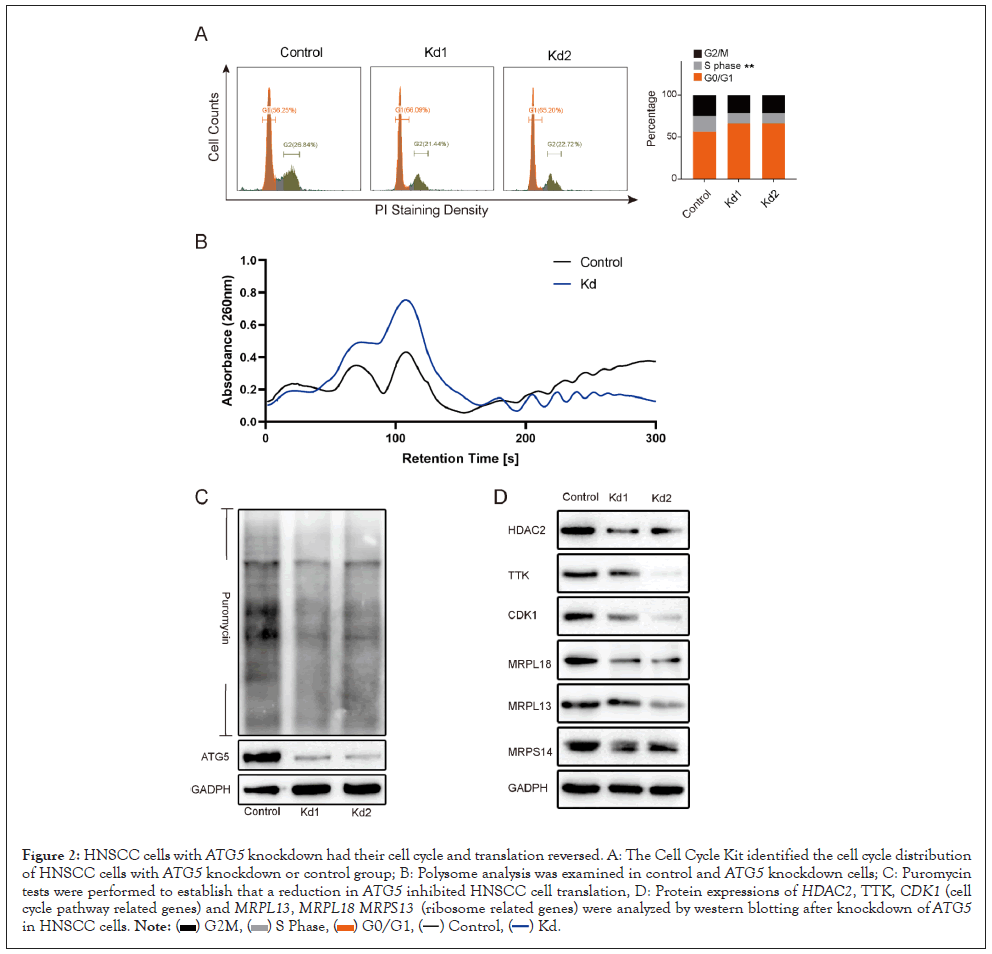
Figure 2: HNSCC cells with ATG5 knockdown had their cell cycle and translation reversed. A: The Cell Cycle Kit identified the cell cycle distribution of HNSCC cells with ATG5 knockdown or control group; B: Polysome analysis was examined in control and ATG5 knockdown cells; C: Puromycin tests were performed to establish that a reduction in ATG5 inhibited HNSCC cell translation, D: Protein expressions of HDAC2, TTK, CDK1 (cell cycle pathway related genes) and MRPL13, MRPL18 MRPS13 (ribosome related genes) were analyzed by western blotting after knockdown of ATG5 in HNSCC cells.
Autophagy is a process in which cellular material is transferred to lysosomes for degrading, allowing for basic cell component turnover as well as the provision of energy and macromolecular precursors [16]. In cancer, autophagy has conflicting, context-dependent functions, and cancer treatments that both activate and inhibit autophagy have been proposed. For instance, autophagy may induce or inhibit apoptosis in the same tumor cell population in various biological settings in exposure to death signals like CD95 Ligand (CD95L) or Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), both of which function as death receptor agonists. Autophagy has been proven in several studies to have a significant role in cancer cell survival. It has been suggested that autophagy contributes to cancer aggressiveness by aiding metastasis. Furthermore, autophagy, as a cellular defense mechanism, has the potential to impair the effectiveness of most chemotherapeutic drugs [17].
Aberrant activation of the central cell-cycle machinery may be found in almost all cancer types and is thought to be a driving force underlying carcinogenesis. According to recent research, cell-cycle proteins govern a wide range of biological activities in addition to stimulating cell division. In fact, genetic defects in the central cell-cycle machinery that cause hyper activation play a role in the formation of almost all cancer types. With the success of CDK4/6 inhibitors in the clinic, it's becoming evident that focusing on certain cell-cycle components might be an effective anti-cancer strategy [18].
Deregulation of protein biosynthesis may also play a role in various facets of malignant tumor, including gene expression, cell signaling, and driving cell biological responses, all of which contribute to cancer development, invasion, and metastasis [19]. The study of translational control's molecular processes may aid in the development of more effective anti-cancer medications and innovative therapeutic prospects. Researchers have recently concentrated on targeting translational machinery to treat cancer, and several tiny molecular inhibitors targeting translation factors or pathways have been evaluated in clinical trials and shown to improve outcomes in a variety of cancer types [20]. Without a doubt, understanding the class of translation regulatory proteins would give a new target for pharmacologic intervention and open new avenues for developing effective anti-tumor therapeutics.
In our study, we investigated the prognosis of the ARG hallmark in HNSCC patients. ATG5 and ATG12 were all shown to be unfavorable prognostic indicators according to the TCGA data. One of the most important regulators of autophagy is ATG5. HNSCC patients with high ATG5 expression had a lower survival rate regardless of Tumor stage, grade, or clinical features. The high ATG5 group's KM curve, on the other hand, was much lower than the low ATG5 groups. Our data revealed that ATG5 was a key factor in HNSCC's poor prognosis. ATG5 overexpression has also been linked to worse clinical outcomes in other malignancies. Positive ATG5 expression, on the other hand, indicates a better prognosis in individuals with breast cancer and osteosarcoma [21].
Atg5-mediated autophagy deficit in the proximal tubules enhances cell cycle G2/M arrest and renal fibrosis, according to researchers [22]. To present, however, the impact of ATG5 on cell cycle, translation, and prognosis in HNSCC remains unclear. In HNSCC, we investigated the correlation between ATG5 and cell cycle and translation-related gene signatures. The influence of ATG5 on cell cycle and translation efficiency was also investigated using HNSCC cell lines.
ATG5 knockdown could inhibit malignant phenotype of HNSCC by reversing cell cycle and translation. HDAC2, TTK, and CDK1 might be one of the processes governing cell cycle control in ATG5-dependent manners. Possible mechanisms involved in ATG5-dependent translation control include MRPL18, MRPL13, and MRPS14. However, function gain and elimination of these genes were needed to be conducted in our further mechanism exploration.
ATG5 is implicated in the cell cycle process and translation regulation in HNSCC and its expression level may impact patient survival, suggesting that ATG5 might be a potentially strong therapeutic target for HNSCC.
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
Citation: Fan H, Weng J, Liu H, Zhang H, Tang S (2022) ATG5 Promotes Head and Neck Squamous Cell Carcinoma by Regulating Translation and Cell Cycle. Immunotherapy (Los Angel). 8:200.
Received: 29-Jun-2022, Manuscript No. IMT-22-17642; Editor assigned: 01-Jul-2022, Pre QC No. IMT-22-17642 (PQ); Reviewed: 15-Jul-2022, QC No. IMT-22-17642; Revised: 22-Jul-2022, Manuscript No. IMT-22-17642 (R); Published: 05-Aug-2022 , DOI: 10.35248/2471-9552.22.8.200
Copyright: © 2022 Fan H, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.