Journal of Hepatology and Gastrointestinal disorders
Open Access
ISSN: 2475-3181
ISSN: 2475-3181
Research Article - (2022)Volume 8, Issue 4
The Hepatitis C Virus (HCV) has a wide geographical and genotype variation within a population all over the world. This research was conducted to identify the genotype distribution of Hepatitis C Virus and its association to the viral load in blood samples. Our goals are to manage the financial burden, promote optimal outcomes, and aid in the personalization of antiviral therapy for Hepatitis C Virus (HCV) treatment. Very few researches have been conducted in Nepal in order to figure out the distribution of various Hepatitis C Virus genotypes in last 25 years. We took up this study to find out the prevalence of various genotypes and subtypes of Hepatitis C Virus and its association to viral load in the patients with chronic Hepatitis C infection in Nepal. Along with the viral load, the HCV genotypes were divided into its seven genotypes and subtypes. By comparing the obtained amplification results, the genotypes were determined. Out of the 100 patients, genotype 3a was accounted for the highest number of cases and found in 53 (53%) patients. Genotype 1a and 1b were found in 23 (23%) and 18 (18%) patients respectively. For the first time, genotype 5a and mixed were identified in Nepalese patients. Genotype 5a was found in 3 (3%) patients. Genotype 6 was found in only 1 (1%) patient. Infection of mixed genotype was discovered in 2 patients. In addition, patients with genotype 3a frequently had higher HCV viral loads compared to those with genotypes 1a and 1b
Hepatitis C virus; Hepatitis C genotypes; Patients; Viral load
A leading global cause of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma is the Hepatitis C Virus (HCV). Infection with Hepatitis C Virus (HCV) has also been identified as the major cause of post transfusion non-A, non-B hepatitis in Nepal. HCV is enveloped, small circular, positive sense and single stranded Ribonucleic Acid (RNA) virus from the genus Hepacivirus, a family Flaviviridae with a diameter of 50 nm and total length of RNA genome is about 9.6 kb [1,2].
HCV strains are classified into seven genotypes (1-7) on the basis of phylogenetic and sequential analysis of whole viral genomes. HCV strains belonging to different genotypes differ at 30%-35% nucleotide sites. HCV is further divided into 67 confirmed and 20 provisional subtypes within each genotype. Six main HCV genotypes (1-6) and numerous distinct subtypes (denoted by a small English alphabet suffixed after genotype, e.g., 1b, 3a, etc.) have been identified thus far based on sequence homology.
In Nepal, HCV Subtype 3b is found exclusively in the eastern region where 25.8% of all HCV-GT3 infections were caused by subtype 3b, and the remaining 74.2% were caused by subtype 3a. In the other two regions, all HCV-GT3 viruses were subtype 3a, and one unidentified GT3 subtype was discovered in the central region. The phylogenetic tree of the 5 untranslated region of the HCV isolates showed a genotype-specific clustering within samples and with reference samples from isolates of India, China and Thailand.
To date, seven genotypes (1-7) of the HCV genome have been defined but most people infected with the virus are unaware of their infection and, for many who have been diagnosed, treatment remains still unavailable though it can be completely cured by treatment with the introduction of new medicines. This is a significant problem because if the disease goes untreated, one third of those who become chronically infected will develop liver cirrhosis and hepatocellular carcinoma. Some studies with small sample size have shown that genotype 1 is found to be comparatively easier for treatment, but despite being a low prevalence area, Nepal has a diversity of hepatitis C and very few researches have been performed on hepatitis C genotypes. Therefore, still a detailed study of genotype distribution in Nepal is needed towards more significant planning for HCV treatment and for and effectively monitoring it. This study was done to determine the change in prevalence of the HCV virus in past few decades, which tends to be increased greatly in very short period of time. Due to the high prevalence and increasing rate of HCV infection day by day, genotyping is more significant for planning the HCV treatment period and effective monitoring the treatment.
This was a prospective type of study. The research was conducted in Decode Genomics and Research Center, Lab, Kathmandu, Nepal. Sampling was taken from the Anti HCV antibody positive patients visiting Decode Genomics and Research Center (DGRC). 100 Samples were taken for this study. Total volume of 4 ml blood sample was collected on Ethylenediamine Tetraacetic Acid (EDTA) vial by following the standard procedure of blood collection. Viral RNA from blood samples was extracted using viral nucleic acid extraction kit (SpinStar™) following manufacturer protocol. Viral load estimation on each blood samples was done using HCV RealStar® viral load estimation kit. Hepatitis C genotype was determined by real time polymerase chain reaction using SpinStar™ HCV genotyping Kit. Data analysis was performed using statistical analysis software SPSS version 20.1. P values <0.05 were regarded as statistically significant.
This study was conducted in Decode Genomics and Research Centre (DGRC), Lab, Kathmandu. The samples were taken and analyzed, for the determination of viral load and genotyping, from the HCV positive patients who visited the DGRC. A total of 100 blood samples were analyzed within four months, of which, 91 were males and 9 were females.
According to WHO guidelines, viral load is classified into two categories, based on its levels. A count less than 8. 0*105 IU/ml is low viral load and a count of more than 8.0*105 IU/ml is high viral load. This quantification was carried out in all 100 HCV positive patients and, the comparison was made between the four groups of HCV genotypes which were detected.
Age wise distribution pattern of HCV patients
The bar graph shown provides information about the age wise distribution pattern of 100 clinically significant HCV positive patients shows in Figure 1. The working age group people have highest rate of HCV infection while the children and old age people have low rate of infection. Out of 100 clinically significant HCV positive patients, 42 belongs to the age group of 31-40, 25 belongs to the age group of 21-40, 20 belongs to the age group of 41-50, 7 belongs to the age group of 51-60 and 4 belongs to the age group of 60-70.
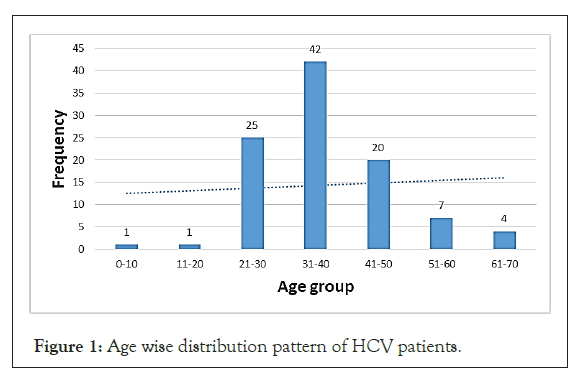
Figure 1: Age wise distribution pattern of HCV patients.
Gender wise distribution pattern of HCV patients
The pie chart illustrates the distribution pattern of male and female HCV infected population shown in Figure 2. Out of 100 HCV patients, 91 were male and 9 were female. Higher number of males is infected with HCV in comparison to female. From this data, we can conclude that male population is at higher risk to HCV infection as compared to female population.
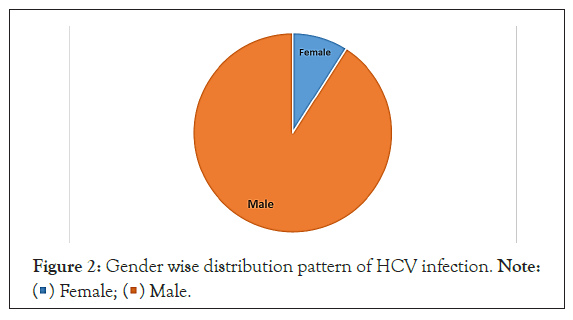
Figure 2: Gender wise distribution pattern of HCV infection. Note:  Female;
Female;  > Male.
> Male.
Prevalence of HCV genotypes in Nepal
The bar graph provides the information regarding distribution of HCV genotypes in 100 HCV infected people. The predominant genotype was 3a (53%), followed by genotype 1a (23%), 1b (18%), 5a (3%) and 6 (1%) respectively. Mixed genotypes were seen in two patients. For the first time, genotype 5a and mixed were identified in Nepalese patients. The genotypes 2 and 4 genotypes were not detected and it is shown in Figure 3.
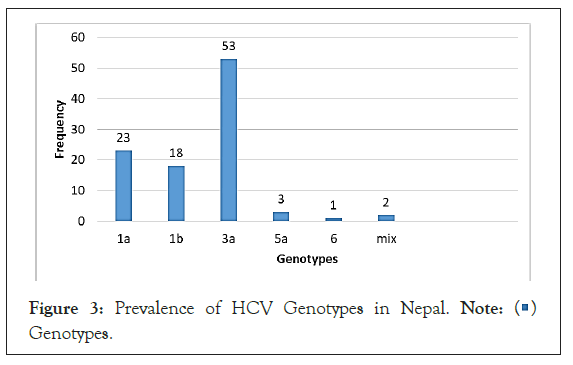
Figure 3: Prevalence of HCV Genotypes in Nepal. Note:  Genotypes.
Genotypes.
Prevalence of HCV genotypes in different age groups
The frequency of HCV genotypes between different age groups. Subtype 1a and 1b were more common in patients of the age groups 21 to 40 years. The frequencies of the genotype 3a subtype were higher in patients of the age group 31 to 40 years. The mixed subtypes 1b, 2 and 1b, 3a were only observed in the age group 31 to 40 and 41 to 50 years as shows in Table 1.
| Age | Genotype | |||||||
|---|---|---|---|---|---|---|---|---|
| 1a Count | 1b Count | 2 Count | 3a Count | 4 Count | 5a Count | 6 Count | mixed Count | |
| <20 | 2 | 0 | 0 | 1 | 0 | 1 | 0 | 0 |
| 20-40 | 15 | 13 | 0 | 37 | 0 | 1 | 0 | 1 |
| 41-60 | 5 | 3 | 0 | 14 | 0 | 1 | 1 | 1 |
| >61 | 1 | 2 | 0 | 1 | 0 | 0 | 0 | 0 |
Table 1: Prevalence of HCV genotypes in different age groups.
Gender wise distribution of HCV genotype
The gender wise distribution of HCV genotype. Out of 91 males included in our study, the predominant genotype was 3a (48/92), followed by genotype 1a (19/91) and 1b (18/91) respectively. Genotypes 5a and 6 were less common in males. 2 mixed infections were also seen in males. On the other hand, out of 9 females, 5 were infected with genotype 3a (5/9) followed by genotype 1a (4/9) shows in Table 2.
| Gender | 1a | 1b | 3a | 5a | 6 | Mixed |
|---|---|---|---|---|---|---|
| Male | 19 | 18 | 48 | 3 | 1 | 2 |
| Female | 4 | - | 5 | - | - | - |
Table 2: Gender wise distribution of HCV genotype.
Distribution of HCV viral load in different genotypes
Viral load less than 2,000 IU/ml is considered to be low, viral load between 2,000 to 20,000 IU/ml is moderate and viral load more than 20,000 IU/ml is considered to be high. The people infected with HCV genotype 3a have a high viral load followed by the people infected with genotype 1a. Accordingly, all the patients infected with 5a have high viral loads. The only case of genotype 6 has moderate viral load, whereas both the cases of mixed genotype have high viral loads is shown in the Table 3.
| Viral load | Genotypes | |||||
|---|---|---|---|---|---|---|
| 1a | 1b | 3a | 5a | 6 | Mixed | |
| Low | 1 | 2 | 2 | 0 | 0 | 0 |
| Moderate | 2 | 4 | 1 | 0 | 1 | 0 |
| High | 20 | 12 | 50 | 3 | 0 | 2 |
Table 3: Distribution of HCV viral load and genotypes.
The p value is less than 0.05, there is significant association between the viral load and genotype as shown in Table 4.
Value |
DF |
Symptoms and Signs. (2-sided) |
|
|---|---|---|---|
Pearson chi-square |
21.995 |
10 |
0.015 |
Likelihood ratio |
15.541 |
10 |
0.114 |
Linear-by-Linear Association |
1.318 |
1 |
0.251 |
N of Valid Cases |
100 |
- |
- |
Table 4: Chi-square tests
Distribution of viral load in different age groups
61% of the patients with high viral load belong to the age group 20- 40 and 19% belong to the age group 41-60. Accordingly, 4% belong to the age group more than 60 and 3% belong to the age group less than 20 shown in Table 5.
| Age | Viral load | ||
|---|---|---|---|
| Low count | Moderate count | High count | |
| <20 | 0 | 1 | 3 |
| 20-40 | 2 | 4 | 61 |
| 41-60 | 3 | 3 | 19 |
| >61 | 0 | 0 | 4 |
Table 5: Distribution of viral load in different age groups.
There are very few studies reporting the presence of various HCV genotypes in Nepal. Shrestha et al., had done HCV genotyping of 12 HCV RNA samples in sera from patients with chronic liver disease. Genotype 2 was detected in five (42%) and genotype 1 and 5 in one each (8%). Genotyping was not possible in the remaining five sera despite high HCV RNA titers in them all.
Shrestha et al., had also reported that incidence of acute Hepatitis C was low, and in Nepal it was confined to drug addicts. In injecting drug abusers in Nepal it was the commonest chronic viral infection 90% were anti-HCV positive and 60% HCV RNA positive [3-9].
Our analysis of HCV infection in 100 patients demonstrated subtype 3a (53%), as we described previously, it is the most prevalent throughout Nepal followed by genotype 1a (23%) and 1b (18%). Genotype 5a, mixed and 6, on the other hand, are considerably less common (3%, 2% and 3%) respectively. One study revealed that HCV genotype 3 is by far the most common genotype accounting for nearly 72% of all HCV infection followed by genotype 1 accounting for nearly 18%. Genotype 2 was found in 3% and genotype 4 and 5 account for 1% each 12. Contrary to this report, no genotype 2 and 4 were detected in our study. The earlier studies as well as the present study indicated that genotype 3 was the most prevalent genotype in central Nepal.
Nepal in 2016 reported that 100 out of the 2700 HIV patients were infected with HCV in sukraraj tropical and infectious disease hospital/teku hospital in Kathmandu, Nepal. The result showed that there was a low frequency of HCV in all the patients infected with HIV. This suggested that the preventive measures for this disease can improve the situation significantly [10-15].
To conclude, different genotypes have been identified in our study. The results from our study highlighted that genotype 3a is the predominant genotype in Nepal followed by genotype 1a and 1b. The severity of liver disease was more in genotype 3a as assessed by higher viral load. Despite being a low prevalence area, Nepal has a diversity of hepatitis C genotype.
The authors declare that they have no Conflict of Interests.
Citation: Pandey G, Gyawali M, Dhungana NS, Paudel R, Dahal S (2022) Distribution of Hepatitis C Genotypes and Viral Load Pattern Among Hepatitis C Positive Patients of Different Age-Group in Nepal. J Hepatol Gastroint Dis. 8:212.
Received: 23-Jun-2022, Manuscript No. JHGD-22-18139; Editor assigned: 26-Jun-2022, Pre QC No. JHGD-22-18139; Reviewed: 11-Jul-2022, QC No. JHGD-22-18139; Revised: 17-Jul-2022, Manuscript No. JHGD-22-18139; Published: 26-Jul-2022 , DOI: 10.35248/2475-3181.22.8.212
Copyright: © 2022 Pandey G, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.