Drug Designing: Open Access
Open Access
ISSN: 2169-0138
ISSN: 2169-0138
Research Article - (2017) Volume 6, Issue 2
This project utilised Verapamil as a lead molecule for iterative design of novel anti-cancer drugs with the potential to inhibit P-Glycoprotein (P-gp), a target which is overexpressed in malignant cells. The protein data bank crystallographic deposition 4M2S, describing the bound co-ordinates of P-gp bound to the small antagonist molecule QZ59-RRR, was used as a template for this study. Two drug design approaches were employed- de novo design and virtual screening (VS). For the former, the SYBYL®-X v1.1 software was used to generate the apo-receptor (empty receptor) and QZ59-RRR extract. Both chiral forms of Verapamil, R and S, were sketched and docked into the P-gp receptor using SYBYL®-X v1.1. 20 optimal conformations were identified and those possessing the best combination of high ligand binding affinity (pKd) and low ligand binding energy (Kcal mol-1) were selected for further analysis. For each of the latter, seed structures were generated based on structure activity relationships (SAR) identified from literature and Poseview®. LigBuilder® v1.2 was used to generate 560 de novo molecules. In the VS approach, the online database ViCi® hosted at the University of Hamburg found 1000 molecules with similar physicochemical properties to the optimal conformations of Verapamil, which served as query molecules in this approach. These were then docked into the protomol, generated in SYBYL®-X v1.1. 107 of the de novo designed molecules and 988 of the virtual screening molecules satisfied Lipinski's Rules. Of these, 88 of the de novo designed molecules had a pKd higher than the templates and 48 of the molecules identified through VS had a pKd that exceeded an arbitrarily set acceptable value of 6. These molecules were subsequently analysed and used to propose a pharmacophore that could be used as a guide in further design of high affinity modulators of P-gp.
<Keywords: 4M2S; Verapamil; P-Glycoprotein; Cancer
Efflux transporters are responsible for eliminating potentially harmful xenobiotics from cells. One of the most prevalent efflux transporters is P-glycoprotein (P-gp) which is expressed in the adrenal glands, brain, kidney and the liver [1]. This transporter is overexpressed in malignant disease [2]. Its expression results in the active extrusion of chemotherapeutic agents from within the malignant cells, consequently rendering them immune to chemotherapy.
Literature indicates that the ATPase activity of the P-gp transporter is stimulated by drugs such as Verapamil [2]. In doing so, the P-gp can be maintained in an ‘occupied’ state, thus preventing it from being able to eliminate chemotherapeutic agents via efflux transport. The aim of this study therefore was to assess the molecule Verapamil and use it as a scaffold for the de novo generation of novel molecules with enhanced affinity, bioavailability [3], specificity and selectivity for the P-gp transporter. Enhanced selectivity is especially important owing to the fact that this would imply decreased affinity for other receptors, such as calcium channels, to which Verapamil is known to bind. This study also aimed to use virtual screening to identify alternative molecules similar physicochemical and inhibitory properties to Verapamil.
X-Ray crystallographic deposition 4M2S [4] describing holo- P-gp bound to the inhibitor molecule QZ59-RRR. Figure 1 was used as a template for this study. Molecular modelling was carried out using SYBYL®-X v1.1 [5]. The bound ligand was extracted from the P-gp Ligand Binding Pocket (LBP) to obtain the apo-receptor. The apo-P-gp and the extracted ligand were then saved in PDB (protein data bank) and mol2 (molecular file) format, respectively.
The 3D structure of Verapamil was sketched, optimised and saved in mol2 format in SYBYL®-X v1.1 [5]. Optimisation of the molecules involved the application of rough dynamic modifications, where the molecule was adjusted in such a way so as to apply the necessary torsions to the sketched molecule to make it as stable as possible. No information was found in the literature [2] regarding which chiral form of Verapamil was responsible for antagonising P-gp and consequently, both possible enantiomers, R and S, were drawn.
Conformational analysis was then carried out using both R- and S- Verapamil (Figures 2 and 3). The bioactive coordinates of the small antagonist molecule QZ59-RRR, which was co-crystallised with the P-gp as described in the template PDB crystallographic deposition 4M2S [4], were used as a docking template. 20 optimal conformations were identified for each enantiomer and individually saved in mol2 format [5].
Figure 2: Verapamil-R conformational results. Rendered with UCSF Chimera® v1.10.1 [11].
Figure 3: Verapamil-S conformational results. Rendered with UCSF Chimera® v1.10.1 [11].
The apo-P-gp receptor and the newly identified 20 optimal conformations of each of the 2 enantiomers of Verapamil were then exported to the Unix-based programme XSCORE_v1.3 [6]. XSCORE_v1.3 [6] was used to calculate the Ligand Binding Affinity (LBA) (pKd) of all of the identified Verapamil conformations at the P-gp binding site. The Ligand Binding Energy (LBE - Kcal mol-1), was also calculated in SYBYL®-X v1.1 [5]. The conformations which combined the highest LBA (pKd) and the lowest LBE (Kcal mol-1) were identified as high affinity molecules which also were energetically stable. These conformations were selected for further analysis. These were Verapamil-R conformation 15, Verapamil-S conformation 4 and Verapamil-S conformation 7, as can be viewed in Graphs 1 and 2. 2 conformations were selected for Verapamil-S as they displayed very similar LBA and LBE values.
Method 1: De novo drug design approach
Seed fragments were modelled in SYBYL®-X v1.1 [5] based on Structure Activity Relationship (SAR) data obtained from the literature [2] and through the generation of 2D topology maps using the software PoseView® [7] (Figures 4-6). A total of 5 seed structures were generated (Figures 7-9).
Figure 4: Structure activity relationship between Verapamil-R conformations 15 with the P-gp active site. The dotted lines represent direct interaction types, whilst the whole lines represent indirect interactions. Both interaction types are labelled with the respective amino acid that they are interacting with. Image rendered with Pose view [10].
Figure 5: Structure activity relationship between Verapamil-S conformations 4 with the P-gp active site. The dotted lines represent direct interaction types, whilst the whole lines represent indirect interactions. Both interaction types are labelled with the respective amino acid that they are interacting with. Image rendered with Pose view [10].
Figure 6: Structure activity relationship between Verapamil-S conformations 7 with the P-gp active site. The dotted lines represent direct interaction types, whilst the whole lines represent indirect interactions. Both interaction types are labelled with the respective amino acid that they are interacting with. Image rendered with Pose view [10].
Figure 7: Showing the seed generated from Verapamil-R conformation 15. The H.spc centre is highlighted in green, carbon atoms in light brown and oxygen in red. Images rendered with UCSF Chimera® v1.10.1 [11].
Figure 8: Showing the seed generated from Verapamil-S conformation 4. The H.spc centre is highlighted in green, carbon atoms in light brown, oxygen in red and nitrogen in blue. Images rendered with UCSF Chimera® v1.10.1 [11].
Figure 9: Showing the seed generated from Verapamil-S conformation 7. The H.spc centre is highlighted in green, carbon atoms in light brown, oxygen in red and nitrogen in blue. Images rendered with UCSF Chimera® v1.10.1 [11].
The P-gp LBP as circumscribed by the selected optimal conformers of Verapamil was analysed in LigBuilder v1.2 [8], thus enabling elucidation of the 3D volume available for novel molecule design, and of a general pharmacophoric structure, designated according to polarity, that all de novo designed ligands would necessarily need to conform to. The designed seed structures were allowed user directed molecular growth at pre-designated H.spc hydrogen atoms using the LINK algorithm of LigBuilder v1.2 [8].
At the end of this process, INDEX files were generated containing all of the de novo generated molecules pertaining to each seed segregated into pharmacophorically similar families. Physicochemical information including chemical formula, molecular weight, LogP, LBA (pKd) and chemical score were also included as descriptors for each molecule.
This process allowed for the identification of family specific pharmacophores and for the identification of the moieties that contribute most significantly to ligand stabilisation and binding.
Method 2: Virtual screening (VS) approach
VS was then used to identify molecular cohorts with binding properties analogous to Verapamil. 2 conformations of Verapamil were selected as queries, one from each enantiomer. These were those, as previously stated, that had the optimal combination of LBA (pKd) and LBE (Kcal mol-1). Specifically, these were Verapamil-R conformation 15 and Verapamil-S conformation 4. The online database ViCi® [9] hosted at the University of Hamburg was used for the VS. Hit molecules were considered similar to the queries from molecular size, shape, topology and outer electronic structure perspectives. MONA® [10,11] was then used to visualise the generated molecules in 2D. These molecules were then filtered to ensure Lipinski Rule [12] compliance and the resulting molecules were inserted into a protomol which was generated in SYBYL®-X v1.1 [5]. This was done to establish a score for each of the molecules so as to identify the optimal cohorts.
During the conformational analysis process, two graphs were plotted, one for Verapamil-R and one for Verapamil-S, showing the LBA (pKd) and LBE (Kcal mol-1) of all of the conformations generated (Graphs 1 and 2). The best conformations from each graph were selected as a scaffold for this study. A total of 107 Lipinski Rule [12] compliant molecules were generated from the 2 molecules. The LBA (pKd) values for the template molecules were 5.56 for Verapamil-R conformation 15, 5.11 for Verapamil-S conformation 4 and 5.62 for Verapamil-S conformation 7.
Results 1: De novo drug design approach
When the molecules that were designed de novo were analysed, the Lipinski Rule [12] compliant structures were evaluated further with a view to identifying which of the moieties, introduced during the de novo design process, contributed most significantly to high affinity relative to Verapamil. This was done in order to identify the key structural differences that contributed most significantly to affinity.
The main structural innovations for the de novo designed molecules were identified. In the case of the molecules that were designed from seed 1 of Verapamil-R conformation 15 (Figure 7) the most important structural innovations were at the level of a connecting chain between the 2 parts of the seed involving hydrogen bond donors (Oxygen). The parts of the original seed were modified in order to increase molecular hydrophobicity. To this end, a number of O-C moieties were substituted with C-C moieties and simplified in a way which minimised the number and length of moiety extensions on the aromatic rings. Table 1 show the optimal molecule designed from this seed structure, belonging to family 13, together with all of its structural modifications and active site interactions.
| Family | Seed 1 Verapamil-R Conformation 15 de novo Molecule | |
|---|---|---|
| 3D Structures | Additional Atoms Added | |
| 13 | 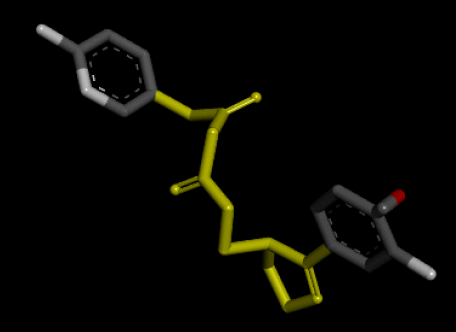 |
Result_109: Addition of 10 Carbons and 3 Oxygens (Yellow). Removal of 2 CH3 moieties and an O-C moiety (White). |
| Result_109 pKd=7.58 |
 |
|
| Number of H-Bonds=4 | ||
| Number of Hydrophobic Interactions=7 | ||
| Number of Unfavourable Interactions=2 | ||
Table 1: Table s howing the de novo molecule Result_109 generated from Seed 1 Verapamil-R Conformation 15 overlapping with the original seed and its subsequent interactions in the active site. Images rendered with Discovery Studio® (Dassault Systèmes BIOVIA).
Novel moieties observed in the de novo molecules originating from seed 1 of Verapamil-S conformation 4 (Figure 8) included a connecting chain between the two parts of the seed which was rich in hydrogen bond donors (Nitrogen and Oxygen) as well as 2 aromatic rings. Parts of the original seed were observed to be modified in such a way so as to increase hydrophobicity. The designed molecules of this particular conformation were also observed to be more compact when compared with the other seeds belonging to the other conformations, with, as observed in Table 2, the replacement of an aromatic ring with an ethyl function. Table 2 shows the optimal molecule designed from this seed, belonging to family 4, with all of its structural modifications and active site interactions.
| Family | Seed 1 Verapamil-S Conformation 4 de novo Molecule | |
|---|---|---|
| 3D Structures | Additional Atoms Added | |
| 4 | 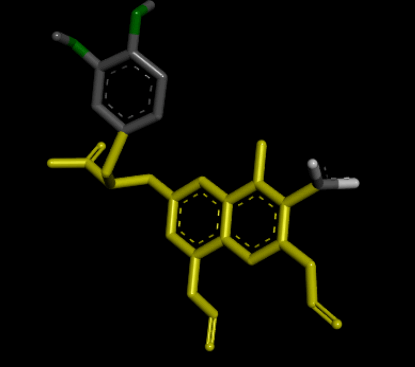 |
Result_32: Addition of 17 Carbons, 3 Nitrogen and 3 Oxygen (Yellow). Substitution of 2 O-C bonds with C-C bonds (Green). Removal of 5 Carbons and 2 Oxygen (White). |
| Result_32 pKd=8.54 |
 |
|
| Number of H-Bonds=6 | ||
| Number of Hydrophobic Interactions=9 | ||
| Number of Unfavourable Interactions=3 | ||
Table 2: Table showing the de novo molecule Result_32 generated from Seed 1 Verapamil-S Conformation 4 overlapping with the original seed and its subsequent interactions in the active site. Images rendered with Discovery Studio® (Dassault Systèmes BIOVIA, 2014).
When the de novo designed structures emanating from seed 1 of Verapamil-S conformation 7 were considered, a connecting chain with a single hydrogen bond donor (Nitrogen) and the substitution of 2 O-C moieties on the original seed structure to 2 N-H bonds was thematic to this molecular cohort. Table 3 shows the highest ranking molecule of this seed, belonging to family 5 with all of its modifications and active site interactions.
| Family | Seed 1 Verapamil-S Conformation 7 de novo Molecule | |
|---|---|---|
| 3D Structures | Additional Atoms Added | |
| 5 | 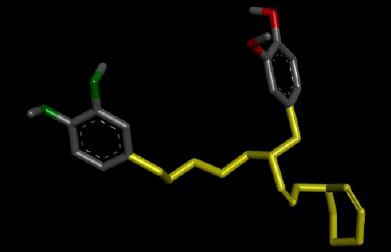 |
Result_19: Addition of 15 Carbons and 1 Nitrogen (Yellow). Substitution of 2 O-C bonds with 2 N-H bonds (Green). |
| Result_19 pKd=6.21 |
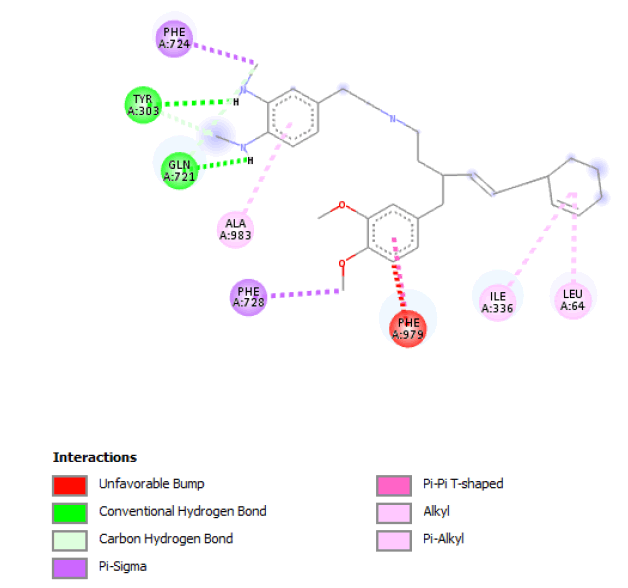 |
|
| Number of H-Bonds=4 | ||
| Number of Hydrophobic Interactions=6 | ||
| Number of Unfavourable Interactions=1 | ||
Table 3: Table showing the de novo molecule Result_19 generated from Seed 1 Verapamil-S Conformation 7 overlapping with the original seed and its subsequent interactions in the active site. Images rendered with Discovery Studio® (Dassault Systèmes BIOVIA).
Modifications noted in de novo molecules formed from seed 2 Verapamil-S conformation 4 include the formation of a connecting chain between the 2 seed parts, which includes hydrogen bond donors (Nitrogen and Oxygen). With regards to the 2 original seed components similar changes as observed in seed 1 of the same conformation were observed, where 2 O-C bonds were substituted with 2 C-C bonds and an aromatic ring was reduced to an ethyl function. Table 4 shows the optimal molecule of this seed, belonging to family 2, with all of its modifications and active site interactions.
| Family | Seed 2 Verapamil-S Conformation 4 de novo Molecule | |
|---|---|---|
| 3D Structures | Additional Atoms Added | |
| 2 | 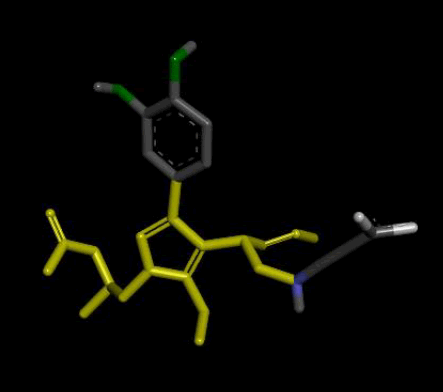 |
Result_101: Addition of 14 Carbons, 2 Oxygen and 3 Nitrogen (Yellow). Substitution of 2 O-C bonds with C-C bonds (Green). Removal of 5 Carbons and 2 Oxygen (White). |
| Result_101 pKd=7.2 |
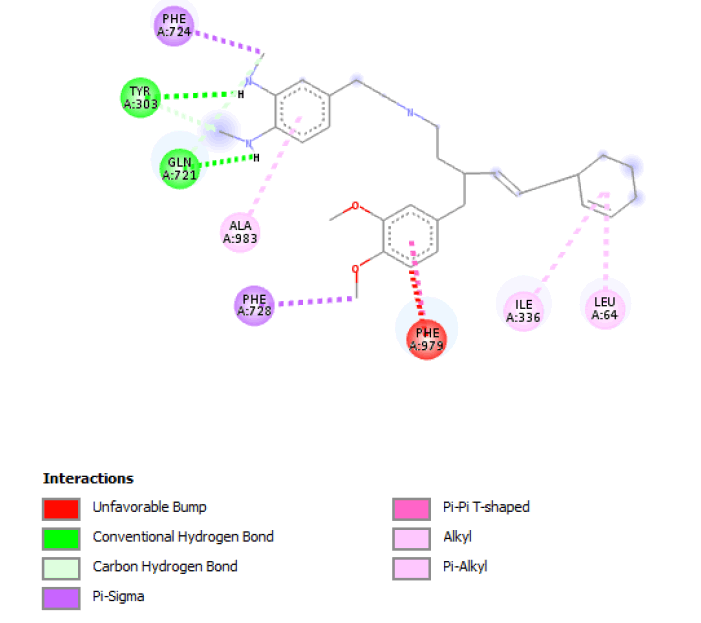 |
|
| Number of H-Bonds=1 | ||
| Number of Hydrophobic Interactions=12 | ||
| Number of Unfavourable Interactions=3 | ||
Table 4: Table sh owing the de novo molecule Result_101 generated from Seed 2 Verapamil-S Conformation 4 overlapping with the original seed and its subsequent interactions in the active site. Images rendered with Discovery Studio® (Dassault Systèmes BIOVIA).
Structural modifications observed in the de novo molecules sourced from seed 2 of the Verapamil-S conformation 7 included the addition of a linking chain with hydrogen bond forming moieties (Nitrogen and Oxygen) included within it. With regards to the 2 original seed components similar changes as observed in seed 1 of the same conformation were observed, where 2 O-C bonds were substituted with 2 N-C bonds. Table 5 shows the optimal molecule of this seed, which came from family 4, with all of its modifications and active site interactions.
| Family | Seed 2 Verapamil-S Conformation 7 de novo Molecule | |
|---|---|---|
| 3D Structures | Additional Atoms Added | |
| 4 | 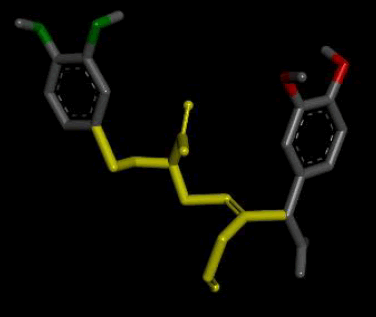 |
Result_98: Addition of 10 Carbons, 1 Oxygen and 1 Nitrogen (Yellow). Substitution of 2 O-C bonds with N-C bonds (Green). |
| Result_98 pKd=6.73 |
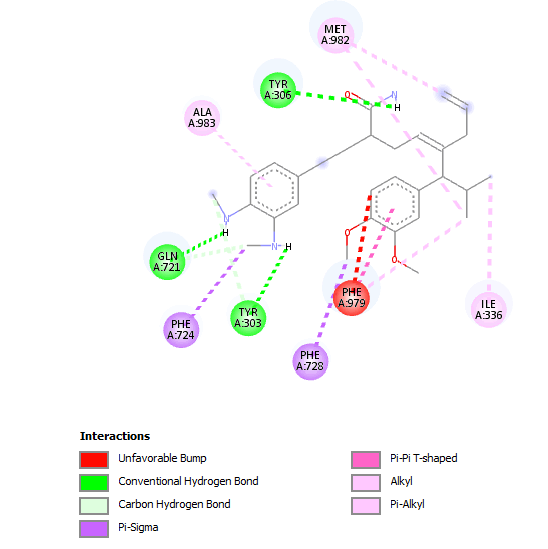 |
|
| Number of H-Bonds=5 | ||
| Number of Hydrophobic Interactions=8 | ||
| Number of Unfavourable Interactions=1 | ||
Table 5: Table showing the de novo molecule Result_98 generated from Seed 2 Verapamil-S Conformation 7 overlapping with the original seed and its subsequent interactions in the active site. Images rendered with Discovery Studio® (Dassault Systèmes BIOVIA).
Examination of the results shows that extensive hydrophobic and hydrogen bond interactions are fundamental for proper binding with the P-gp binding site. Hydrophobic interactions were observed to act primarily on the 2 aromatic rings at the ends of the molecules. Hydrogen bonds were formed on the linking chain and also on the aromatic rings at the ends of the molecules. Key substituents were identified as being important for binding and are highlighted in Table 6.
| Seed from which substituent was obtained | Substituent |
|---|---|
| Seed 1 Verapamil-R_15 | 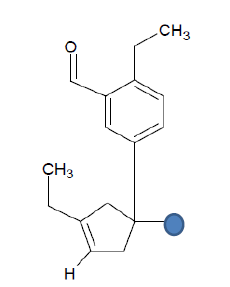 |
| Seed 1 Verapamil S_4 | 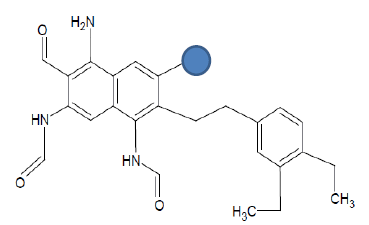 |
| Seed 2 Verapamil S_4 | 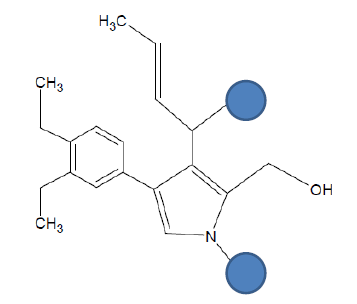 |
| Seed 2 Verapamil S_7 | 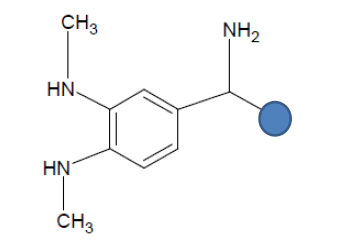 |
Table 6: Table showing the substituents which are essential for binding, as identified the results obtained from 4 of the 5 seeds which provided the best results. Blue points on images represent variable R groups. Images rendered with ChemSketch® v11.02 (ACD/Chemsketch).
Results 2: Virtual screening (VS) approach
When VS was carried out, a total of 1000 molecules was generated for each chosen conformation and then inserted into the generated protomol in order to assess the binding affinity of these molecules for this energetically dissatisfied space. The benefit of using a protomol in this scenario was that it provided a larger pharmacophoric volume for design, owing to the fact that the protomol incorporates a molecular volume larger than the bioactive volume that was elucidated when the optimal conformers of Verapamil were used to probe the P-gp. This is evident in that the volume of the Verapamil circumscribed LBP was 721.5 Å, as compared to that of 1116Å for the protomol.
When the optimal conformation of Verapamil-R was used to query the online molecular database ViCi [9], a total of 500 Lipinski Rule [12] compliant hits were identified, 60 of which had a LBA (pKd) which was higher than that of the query. The optimal molecule had a calculated LBA (pKd) of 7.72 and may be viewed in Table 7.
| Verapamil-R conformation 15 | |
| Chemdiv_F344-0724 | |
| 3D Structure | 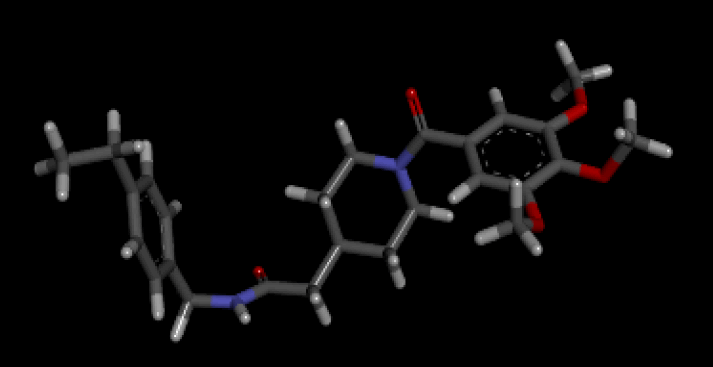 |
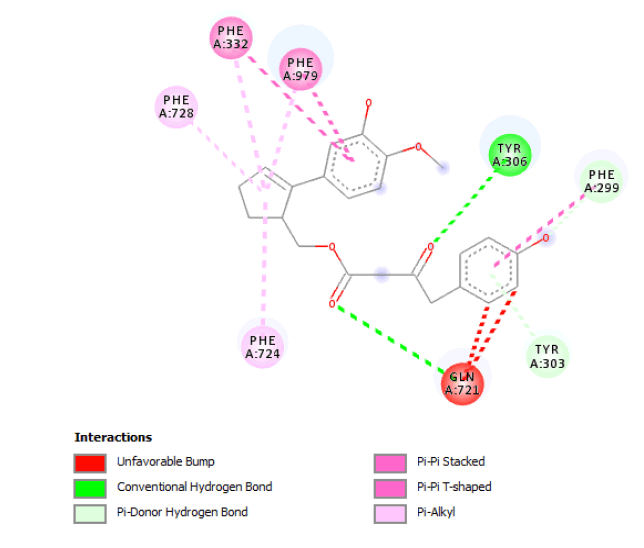
| 2D Structure | 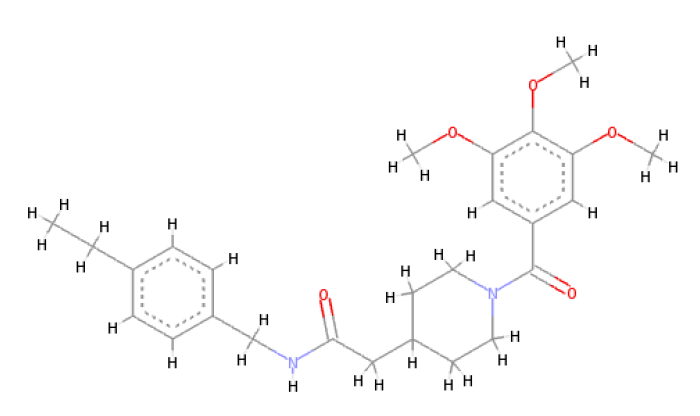 |
Table 7: Showing the 3D and 2D structure of the molecule obtained via virtual screening which had the highest pKd. Images rendered with Discovery Studio® (Dassault Systèmes BIOVIA).
The optimal conformer of Verapamil-S yielded a total of 488 molecules which were Lipinski Rule [12] compliant, 99 of which had a LBA (pKd) which was higher than the original query molecule. The optimal hit molecule was identified as having a LBA (pKd) 7.87 and may be viewed in Table 8.
| Verapamil-S conformation 4 | |
| LC_F5226-0228 | |
| 3D Structure | 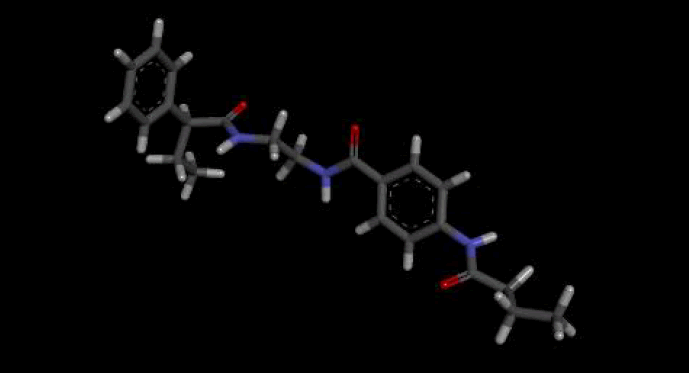 |
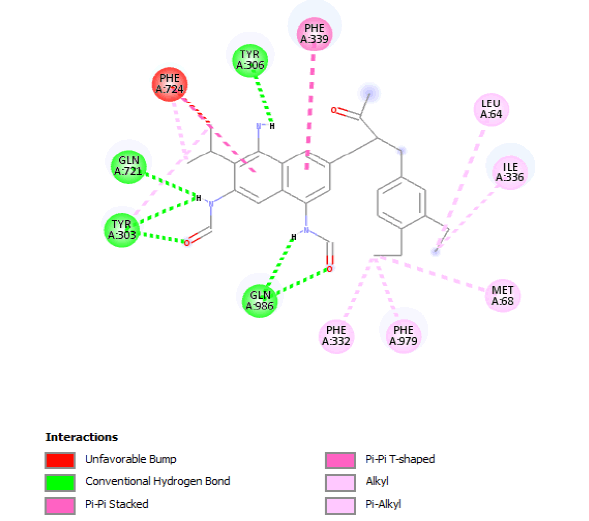
| 2D Structure | 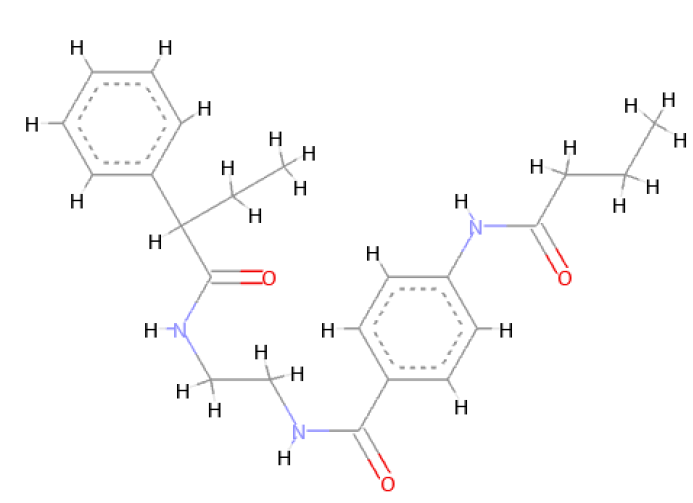 |
Table 8: Showing the 3D and 2D structure of the molecule obtained via virtual screening which had the highest pKd. Images rendered with Discovery Studio® (Dassault Systèmes BIOVIA).
Examination of these molecules show the same distribution of hydrophobicity and hydrogen bond forming moieties, consequently reenforcing the structural innovations observed and discussed in the de novo approach.
Results show that the hydrophobic and hydrogen bond distribution of the molecules are fundamental for binding with the P-glycoprotein active site. Results of other studies, such as those conducted by Ekins et al. [13] show that a minimum of 2 hydrophobic groups, 1 hydrogen bond acceptor group and an aromatic core are required for proper inhibition of the P-glycoprotein receptor. These results have been emulated in this project, where the highest ranking molecules have shown to possess these functional groups.
The study proves to have potential as it has been investigated using 2 different approaches, de novo and virtual screening. The 2 approaches have strengthened the level of assurance in the substituents which have been found to be important for binding, as they have been found to be present in the molecules generated from both approaches.
A major limitation of this study is the static approach that was taken in both de novo and virtual screening approaches and only single bond rotations were allowed when handling ligands and receptors via software. In addition to this, the X-Ray crystallographic structure 4M2S [14] had a resolution of 4.4A, which is relatively poor; alas it was the best crystallographic structure of P-glycoprotein at the time of selection.
Due to time constraints, the SAR interactions of the molecules which were generated via ViCi® Hamburg [9] and LigBuilder® v1.2 [8] were not studied in depth with the use of other software. This could have been beneficial as more molecular possibilities could have been identified which could have optimised binding with the P-glycoprotein site even further.
The de novo and VS drug computational work carried out in this study, helped identify moieties and also structural molecules which could potentially be used for the future development of newer and more effective molecules for the inhibition of P-glycoprotein in cancer cells. The structure activity relationships analysed also help identify molecular features that are fundamental for binding with the active site in a more effective way than the template Verapamil molecule. All of the molecules generated in this study can also be made available to other research projects or online molecular databases to further bolster scientific advancements in the field of drug design. The next step following the computational work would be to finalise drug design and commence the validation process with the conduction of in vitro and in vivo assays.
I would like to thank Dr. Claire Shoemake for her constant help and support.