Journal of Developing Drugs
Open Access
ISSN: 2329-6631
ISSN: 2329-6631
Research Article - (2019)Volume 8, Issue 1
Female Aedes aegypti and Anopheles gambiae mosquitoes are living vectors liable for transmitting many parasitic and dreadful viral diseases. The major target is acetyl cholinesterase (AChE) 1 enzyme for the parasite transmission. Anopheles gambiae carries AgAChE1 enzyme responsible for the malaria parasite whereas Aedes aegypti carries AaAChE1 enzyme that injects dengue, yellow fever, Zika, and chikungunya viruses to the healthy individuals. These vector-borne infections are now a major public threat. Deforestation and global warming may cause drastic change of the climate which causes vector-borne disease to re-emerge worldwide. Control of mosquito vector saves many lives. One of the mechanisms to kill the mosquito vector is to use insecticides such as chlorinated hydrocarbons, organophosphates, carbamates, and pyrethroids recommended by the World Health Organization (WHO). These are covalent inhibitors of acetyl cholinesterase enzyme and their accumulation may produce ecotoxicity to the nontargets including aquatic animals and humans. It was shown that non covalent phenoxyacetamide-based inhibitors can specifically target acetyl cholinesterase (AChE) 1 responsible for the vector transmission. Therefore, an attempt has been made to explore AChE1 target via ligand based quantitative structure-activity relationship (QSAR) modeling on phenoxyacetamide-based compounds to predict crucial features of these inhibitors responsible for the design of highly active ligands.
Aedes aegypti mosquito vector; Computed structural descriptors; GA-MLR based QSAR; AChE1; Phenoxyacetamide
Vector-borne diseases are the infections which are transmitted via living organisms that represent the vector. These vectors are mosquitoes, ticks, flies, sandflies, fleas, triatomine bugs, and some freshwater aquatic snails. The disease-vector bites an infected animal or human and then transmits pathogens through blood meals between vertebrate host animals and humans. Thus the infection spread from an infected fellow to the healthy individuals [1,2]. Vector-borne infections become a pandemic threat because more than one billion people get infected each year, engulfing more than 7,00,000 annual deaths caused by malaria, dengue, schistosomiasis, human African trypanosomiasis, leishmaniasis, Chagas disease, yellow fever, Japanese encephalitis, and onchocerciasis, globally [3]. Since 2014, outbreaks of dengue, malaria, chikungunya, yellow fever and Zika have claimed a strong helmet over the health system protection in many countries. Therefore, control of the mosquito vector is a major target nowadays [3]. Four types of insecticides including chlorinated hydrocarbons, organophosphates, carbamate, and pyrethroids were recommended by the World Health Organization (WHO) for controlling mosquito-vectors [4,5]. These compounds can damage the neurotransmission signaling system. Acetylcholine (Ach) releases into the synaptic cleft where it binds to Ach receptors at the post-synaptic membrane, transferring the signal to downstream nerve cells. Acetyl cholinesterase (AChE) causes breakdown of the Ach to produce choline which is being re-used for further synthesis of Ach to proceed synaptic signaling system. Inhibition of AChE causes an accumulation of acetylcholine, leading to overstimulation and then paralysis of the nervous system [6,7]. Therefore, inhibition of AChE is a major target to control the mosquito vector.
Insecticide resistance and toxicity become a major drawback [8,9]. To quest least toxic chemicals, covalent inhibitors such as carbamate derivatives and difluoromethyl ketones were being developed as inhibitors of AChE1 and AChE2 enzymes responsible for transmission of vector found in Anopheles gambiae (AgAChE1; vector of the malaria parasite) and Aedes aegypti (AaAChE1; vector of the dengue, yellow fever, Zika, and chikungunya viruses). But these compounds failed due to lack of desired selectivity and potency [10-12]. Therefore, much more attention was paid nowadays on to the discovery of non-covalent inhibitors acetylcholinesterase 1 (AChE1) as the role of AChE2 is still not fully understood [13-18].
Alout et al. [19] tested a non-covalent, reversible class of pyrimidinetrion furan-substituted compounds that showed promising potency on AgAChE1 and AgAChE1-G122S both in vitro and on mosquito larvae, however, the selectivity profile of these compounds was not reported. Knutsson and co-workers [20] developed many N-aryl-N'-ethyleneaminothioureas which proved to be inhibitors of AChE1; the most efficient following compound showed sub-micromolar potency. Importantly, the inhibitors exhibited selectivity over the human AChE (hAChE), which is desirable for new insecticides. The structure-activity relationship (SAR) analysis of the thioureas revealed that small changes in the chemical structure had a large effect on inhibition capacity. Knutsson et al. [21] recently discovered a number of non-covalent inhibitors that selectively target Acetylcholinesterase 1 (AChE1) of the mosquitoes An. gambiae and Aedes aegypti (Ae. aegypti, transmitting dengue, chikungunya, and Zika virus infections). AChE1 is a validated insecticide target to control mosquito vectors of e.g., malaria, dengue, and Zika virus infections. The insecticides most commonly used for vector control disrupt the insect’s nervous system by inhibiting voltage-gated ion channels (pyrethroids and organochlorines) or by inhibiting the essential enzyme acetylcholinesterase (AChE) (organophosphates and carbamates). The physiological role of AChE is to terminate nerve signaling by rapidly hydrolyzing the neurotransmitter acetylcholine. The design, synthesis, in vitro and in vivo evaluation of phenoxyacetamide-based analogs were done. The aim was to explore the molecular basis for the inhibitors’ selectivity for mosquito AChE1 versus hAChE and their potency on G122S mutated AgAChE1. In a recent differential high-throughput screen (HTS) studied against the AChE1 enzymes from Anopheles gambiae (An. gambiae; vector of the malaria parasite; AgAChE1) and Aedes aegypti (Ae. aegypti; vector of the dengue, yellow fever, chikungunya, and Zika virus infection; AaAChE1), Engdahl et al. identified a number of non-covalent phenoxyacetamide based hits showing potential selectivity for the mosquito enzymes over the human enzyme [22]. But there is hardly any QSAR formulated on these compounds till date. Therefore, it is our target in the present study to explore structural features for the design of congeneric potent phenoxyacetamide compounds to inhibit AChE1 responsible for the transmission of ZIKV mosquito vector.
Biological activity data
A total number of 23 phenoxyacetamide derivatives (Table 1) showing half maximum inhibitory effect on enzyme acetylcholinesterase 1 (AChE1) of AaAChE1 that represents Aedes aegypti (Ae. aegypti; vector of the dengue, yellow fever, chikungunya, and Zika virus infection). The design, synthesis and in vitro activity evaluation of phenoxyacetamide-based analogs were done to explore the molecular basis for the inhibitors’ selectivity towards mosquito AChE1 [21,22]. These inhibitors target the essential enzyme acetylcholinesterase which terminates cholinergic transmission through rapid hydrolysis of the neurotransmitter acetylcholine.
| Comp No. |
Structure | pIC50 (µM) | Comp No. |
Structure | pIC50 (µM) |
|---|---|---|---|---|---|
| 1. | 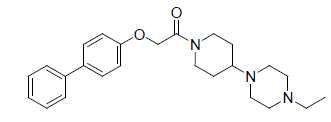 |
0.657 | 2. | 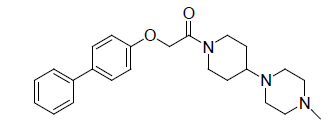 |
0.602 |
| 3. | 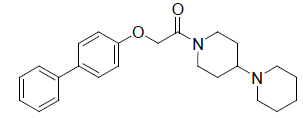 |
-0.602 | 4. | 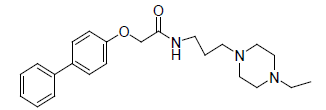 |
-0.518 |
| 5. | 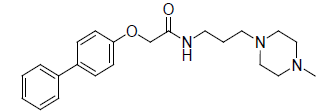 |
-1.079 | 6. | 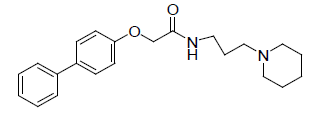 |
-0.995 |
| 7. | 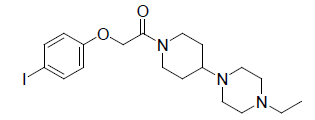 |
-0.176 | 8. | 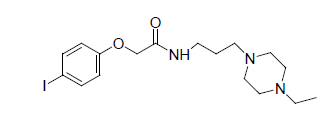 |
-1.857 |
| 9. | 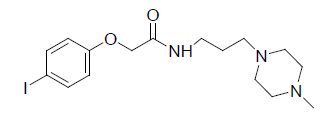 |
-2.049 | 10. | 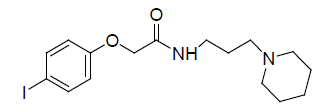 |
-1.556 |
| 11. | 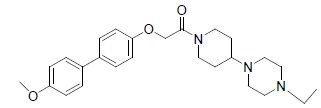 |
0.638 | 12. | 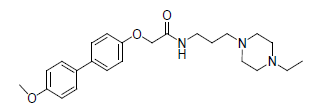 |
-0.579 |
| 13. | 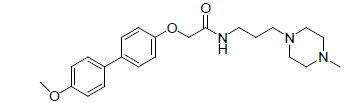 |
-1.204 | 14. | 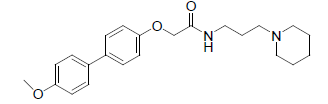 |
-0.591 |
| 15. | 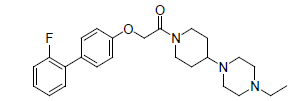 |
0.657 | 16. | 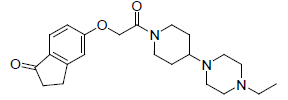 |
0.698 |
| 17. | 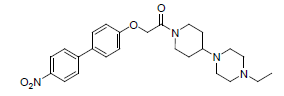 |
0.958 | 18. | 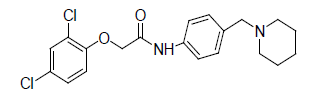 |
0.356 |
| 19. | 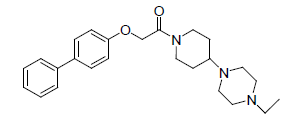 |
0.657 | 20. | 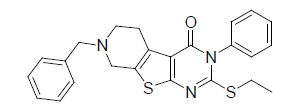 |
-0.944 |
| 21. | 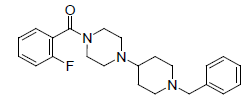 |
-1.079 | 22. | 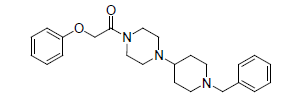 |
-1.278 |
| 23. | 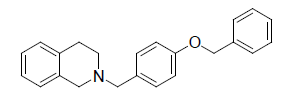 |
-0.462 |
Table 1: Biological activity data.
Structure formulation and calculation of molecular descriptors
All the structures of 23 phenoxyacetamide derivatives were formulated using 2D ChemDraw. The 2D structures were then converted into 3D modules and the geometries of all compounds were fully minimized using MM2 force field considering the default conversion procedure [23]. Fully optimized 3D structures were browsed into PaDEL descriptor which is freely accessible open source software for calculation of theoretical molecular descriptors classification into 1D, 2D, and 3D respectively. Many 1875 structural invariants were calculated using the Chemistry Development Kit based on the code of Java language [24,25].
Theoretical molecular descriptors or the structural invariants encode molecular configuration and properties in terms of physicochemical constitutional, geometrical and three dimensional (3D), electrostatic, functional, atom centered fragments and topological indices, respectively. Biological activity (pIC50) is quantitatively correlated with various structural invariants to formulate QSAR model in terms of statistical regression methods. QSAR based on computed structure descriptors may provide a significant tool for the rational design of potent molecule in theoretical drug design and discovery research [26-31].
Statistical GA-MLR modeling
1875 descriptors were calculated and reduced to 1071. The reduced set of theoretical molecular descriptors (Figure 1) was taken into consideration for the development of a quantitative structureactivity relationship model. Descriptors with perfectly constant and highly inter-correlated descriptors were removed considering variance and correlation coefficient cut-off values of 0.0001 and 0.99 using V-WSP algorithm of Nanobridges software [32]. The reason is that the highly inter-correlated descriptors may contribute chance correlation and produce overfitting of the model. As the number of structural predictors greatly exceeds the number of compounds, the selection of important predictors is necessary for the QSAR modeling. Genetic algorithm-multiple linear regression (GA-MLR) has been used for the development of QSAR model considering reduced predictors data after variable selection by genetic algorithm method [33,34]. The GA methods uses a binary string of digits containing the values of “1” or “0” for presence and absence of the invariant. The length of each string is the same and is equal to the total number of descriptors. Quality of the model is calculated by the fitness function by taking 100 different random combinations of the calculated molecular descriptors.

Figure 1. Descriptors used in current study.
Fitness function of each model is formulated in term of Q2 Loo or R2 where, Q2 Loo represents cross-validated R2. Values of Q2 Loo and R2 are calculated by the standard equation [27,35,36].
Total data set comprise of 23 phenoxyacetamide compounds. A number of training and test set was randomly generated. Many training QSAR models have been developed utilizing various sets of computed molecular descriptors including 1D, 2D, and 3D respectively using GA-MLR methods of NanoBridges software. The impact of the different classes of computed descriptors on the inhibition of acetylcholinesterase1 (AChE1) by phenoxyacetamide compounds has been carried out. It was shown that combination of 1D, 2D and 3D descriptors give highest AChE1 inhibitory impact in terms of good quality model evaluation parameters denoted by R2 (R is the square root of multiple R-square for regression), Q2Loo (cross-validated r2) values for the training set, an external validation was performed by calculating predictive R2 (R2pred) and the standard error of estimation, SEE, and modified r2 (r2m). R2, Q2Loo, R2 pred, and r2 m are calculated by using standard statistical equations [27,35-38]. The value of R2 should be greater than 0.6, whereas Q2Loo, Rpred 2 [37,38] and r2 m which denotes predictive statistics should be greater than 0.5 [39]. Since the data set is small (only 23 compounds), three QSAR models with 3 different test set compounds showing the best results on the AChE1 synthesis inhibition is given in Table 2.
| Model-1 | |
| pIC50=8.168(+/-1.164) -0.001(+/-0.0001) ATSC8m -1.147(+/-0.957) VE1_Dze -19.051(+/-2.633) E2e N=16, R2=0.867, Q2Loo=0.809, R2pred=0.5913, r2m (test)=0.686, SEE=0.323 | |
| Parameters | Physical interpretation |
| ATSC8m | Centered Broto-Moreau autocorrelation of lag 8 weighted by mass |
| VE1_Dze | The coefficient sum of the last eigenvector from Barysz matrix / weighted by Sanderson electronegativities |
| E2e | 2nd component accessibility directional WHIM index / weighted by Sanderson electronegativity |
| Model-2 | |
| pIC50=-3.456(+/-2.773)-0.966(+/-0.299) TDB9r -8.403(+/-0.795) E2v +2.475(+/-0.787) SpMax6_Bhi N=14, R2=0.938, Q2Loo=0.882, R2pred=0.7891, R2m (test)=0.578, SEE=0.211 | |
| Parameters | Physical interpretation |
| TDB9r | 3D Topological distance based descriptors - lag 9 weighted by covalent radius |
| E2v | 2nd component accessibility directional WHIM index / weighted by van der Waals volume |
| SpMax6_Bhi | largest eigenvalue n. 6 of Burden matrix weighted by ionization potential |
| Model-3 | |
| pIC50=1.127(+/-0.518) + 0.101(+/-0.019) RDF90u + 1.447(+/-0.818) E2m -10.835(+/-1.50411) E2v N=17, R2=0.890, Q2Loo=0.803, R2pred=0.946, r2m (test)=0.871, SEE=0.313 | |
| Parameters | Physical interpretation |
| RDF90u | Radial distribution function - 090 / unweighted |
| E2m | 2nd component accessibility directional WHIM index / weighted by mass |
| E2v | 2nd component accessibility directional WHIM index / weighted by van der Waals volume |
Table 2: Best QSAR models.
The training model 1 is based on 70% of total data set whereas model 2 is formulated using 61% of the total data set. The compound numbers 1, 5, 7, 9, 11, 15 and 17 were treated as a test set for model 1 whereas molecule number 1, 8, 12-13 and 15-19 were taken as a test set in case of development of QSAR model 2. Model 3 consists of 74% and 26% as training and test set of the total data. Test set molecules are 1, 4, 6, 8, 10 and 17. So diversity is there for the training and test set generation. R2, Q2Loo, R2 pred, and r2 m for all models 1-3 are acceptable.
Further, the Pearson correlation coefficient between the descriptors which arrived in the best QSAR models 1-3 are given in Table 3. It was shown that chance correlation is zero for the developed models 1-3 because the correlation between the descriptors was calculated as ≤ 0.33 except E2v and E2m which shows 0.602. It is also acceptable.
| Parameters of model 1 | |||
| Parameters | ATSC8m | VE1_DZe | E2 |
| ATSC8m | 1.00 | - | - |
| VE1_DZe | -0.411 | 1.00 | - |
| E2 | -0.117 | 0.330 | 1.00 |
| Parameters of model 2 | |||
| Parameters | TDB9r | E2v | SpMax6_Bhi |
| TDB9r | 1.00 | - | - |
| E2v | -0.159 | 1.00 | - |
| SpMax6_Bhi | -0.167 | -0.545 | 1.00 |
| Model Parameters of model 2 | |||
| Parameters | RDF90u | E2m | E2v |
| RDF90u | 1.00 | - | - |
| E2m | -0.157 | 1.00 | - |
| E2v | -0.123 | 0.602 | 1.00 |
Table 3: Pearson correlation coefficient between the modeled descriptors.
To examine the further predictive power of the models, QSARs 1-3 are used to predict the biological activities of the corresponding test sets. Correlation between observed and predicted activities was graphically plotted in (Figures 2-4). The correlation coefficient (r) between observed vs. predicted activities were calculated as 0.886, 0.892 and 0.990 respectively which stands significant model validation.
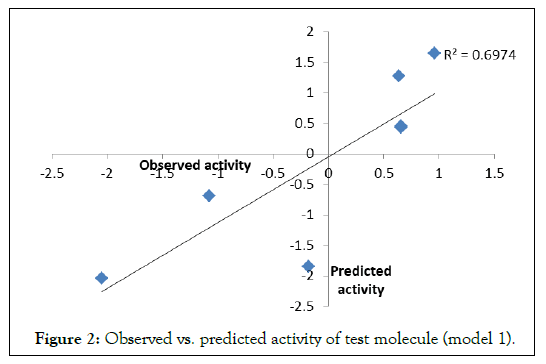
Figure 2. Observed vs. predicted activity of test molecule (model 1).
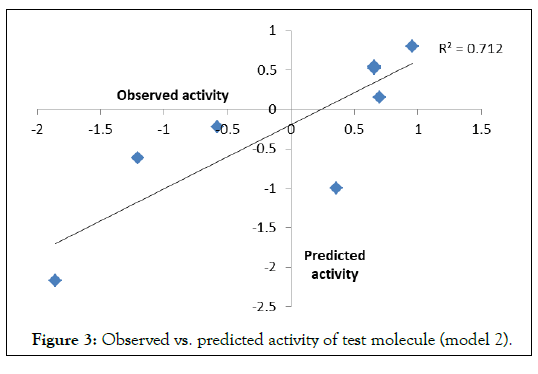
Figure 3. Observed vs. predicted activity of test molecule (model 2).
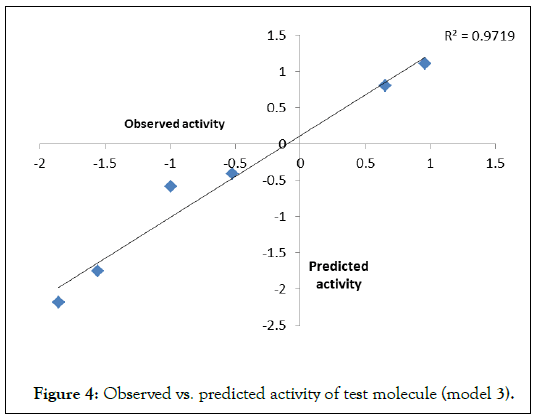
Figure 4. Observed vs. predicted activity of test molecule (model 3).
From the QSAR results, it was shown that ATSC8m, VE1_Dze, E2e, TDB9r, SpMax6_Bhi, RDF90u, E2m, and E2v are important descriptors captured in the training QSAR models 1-3 responsible for producing inhibition of target enzyme AChE1.
QSARs 1-3 depicted that these molecules should produce the ionizing potential which represents electronegativities and electrostatics interaction towards receptors. It was captured by modeled parameters including VE1_Dze, E2e, E2v, SpMax6_Bhi and RDF90u invariants of the phenoxyacetamide. From the above QSAR study, it was predicted that these compounds should not produce covalent linkage towards AChE1. It is denoted by the property TDB9r with negative regression coefficient which focuses on 3D topological radius distance based covalent radius already captured in QSAR model 2.
Out of these three QSAR, model 3 represents the best results. The modeled parameters RDF90u encodes radial distribution function which is correlated with the electronegativities due to electron delocalization. The parameter E2m representing 2nd component accessibility directional WHIM index/weighted by mass encodes the atomic distribution and shape. In this QSAR study, it was also found that the groups having higher van der Waals radius are not favorable. Therefore small electron-withdrawing groups such as Cl, Br, F, and CCl3 responsible for electron delocalization may be favorable to produce potent congeneric phenoxyacetamide compounds having inhibition of AchE1 enzyme. Thus it is an attempt to Control of Aedes aegypti mosquito vector transmitting dengue, yellow fever, chikungunya, and Zika virus infection.
Citation: Naaz A, Kumar M, Sharma A, Teotia D, Nandi S, Saxena M (2019) Control of Aedes aegypti Mosquito Vector by Attempting QSAR Modeling on Phenoxyacetamide-Based Inhibitors to Target AChE1. J Develop Drugs 8:192.
Received: 26-Feb-2019 Accepted: 05-Mar-2019 Published: 12-Mar-2019
Copyright: © 2019 Naaz A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.