
Journal of Clinical Toxicology
Open Access
ISSN: 2161-0495

ISSN: 2161-0495
Research Article - (2022)Volume 12, Issue 4
Acute Hepatopancreatic Necrosis Disease (AHPND) is a bacterial disease of white leg shrimp, which has a high mortality rate (100%) and economic losses. Our objective was to identify the genes which caused cell and organ toxic damage and applied bioproducts to prevent it. Litopenaeus vannamei shrimp were collected from an infected pond in Central Vietnam and analyzed at the Institute of Biotechnology, Hue University. The PirA gene of Vibrio parahaemolyticus strain K5 was isolated and analyzed for nucleotide sequence, paired with the expression vector pQE30. The expression vector was transformed into E. coli strain M15, the PirA recombinant protein was expressed in the form of 6xHis-PirA fusion protein of about 15 kDa. PirA recombinant protein was purified and determined the PirAvp binding ratio, cloning, and sequencing of PirA gene from Vibrio parahaemolyticus strain K5 causing AHPND by PCR method with specific primers and molecular weights of PirAvp and the PirAvp complex. PirA gene from Vibrio parahaemolyticus strain K5 was cloned into a pGEM-T easy vector (Promega, USA) and screened E. coli TOP10 colonies containing pGEM T easy/PirA recombinant plasmid on LB agar/ampicillin/ IPTG/X-Gal medium. PCR showed a band of about 347 bp, matching the size of the PirA gene and two nucleotide sequences (BamHI and HindIII). The results showed that the PirA gene has a length of 336 bp and is like the PirA gene on GenBank (Code: KU556825.1). The results of protein extracted from E. coli M15 recombinant cells and 6xHis-PirA target protein was collected in elution fractions from EF2 to EF6, showed that the concentration of 6xHis-PirA protein and EF3 elution fraction collected the highest protein concentration (1,586.54 µg/ml). The purified PirA recombinant protein will provide materials for development research to create biological products to prevent and treat AHPND.
Vibrio parahaemolyticus; PirA gene; White leg shrimp; K5 caused
Acute Hepatopancreatic Necrosis Disease (AHPND), also known as early mortality syndrome or acute hepatopancreatic necrosis syndrome, is a disease caused by Vibrio strains and affects shrimp. AHPND was first detected in China in 2009 and spread throughout Southeast Asia to countries such as Thailand, Vietnam, Malaysia and the Philippines [1-4]. In 2013, the disease was found in Mexico [5], and in other countries in Latin America between 2013 and 2015 [6]. Outbreaks of AHPND disease have also been reported in Bangladesh [7,8]. Shrimp production in these countries has declined sharply [9]. AHPND is characterized by various clinical signs, including an empty gastrointestinal tract, opaque white stomach, hepatopancreas atrophy, a pale color, lethargic swimming, stopping of eating, and soft shell [2,10]. The disease progresses rapidly, starting about 8 days after stocking, and the highest mortality occurs during the first 20 to 30 days (up to 100%) in Litopenaeus vannamei and Penaeus monodon populations [10,11], causing heavy economic losses of billions of dollars each year to shrimp farmers around the world [12]. Currently, there is no effective preventive measure.
One of AHPND pathogens has been identified as a specific strain of Vibrio parahaemolyticus [10,13] recorded the presence of V. parahaemolyticus and Vibrio alginolyticus with a very high density of 13.106 CFU/mg from the hepatopancreas mass of white leg shrimps in Thua Thien Hue (Vietnam) with clinical signs of AHPND [10,12] identified some characteristics of Vibrio parahaemolyticus strain V1 isolated from juvenile white leg shrimp showing clinical signs of AHPND in Thua Thien Hue. The study revealed it is a Gram-negative bacterium, rod shaped, 0.3-0.5 µm in width and 1.4-2.6 µm in length. In addition, its colonies are mauve, round, smooth with a darker center than the surrounding on CHROM agar medium [12]. V. parahaemolyticus is mainly distributed in marine and estuarine environments around the world [14]. The strains of AHPND pathogenic bacteria contain an intracellular plasmid, which is not found in non-pathogenic strains [15]. This large plasmid (69-70 kb) contains PirAvp and PirBvp toxin genes [16,17], that encode PirABvp binary protein, which has been shown to be the major virulence factor [16,18-20] isolated 14 strains of Vibrio spp. carrying PirAvp and PirBvp genes from white leg shrimp samples cultured in Tam Giang lagoon [20]. From white leg shrimps infected AHPND in Phong Dien district, Thua Thien Hue province (Vietnam), we isolated V. parahaemolyticus strain K5 carrying both PirA and PirB toxin genes with predicted size of 336 bp and 1,317 bp, respectively.
In this study, we sequenced the PirA gene of the isolated V. parahaemolyticus strain K5, created E. coli M15 recombinant cells carrying pQE30/PirA recombinant plasmid as well as induced the expression of PirA recombinant protein in the form of 6xHis- PirA fusion protein. Then, the recombinant antigen is purified to provide materials for further research to develop vaccines or antibodies against AHPND.
Materials
E. coli strain TOP10 (Promega, USA), E. coli strain M15 (Qiagen, Germany), pGEM-T-Easy vector (Promega, USA), pQE30 expression vector (Qiagen, Germany), PirA gene isolated from Vibrio parahaemolyticus strain K5 [21].
Chemicals: Tryptone (Biobasic, USA), peptone (Biobasic, USA), yeast extract (Biobasic, USA), NaCl (Merck, Germany), KCl (Merck, Germany), MgCl2 (Merck, Germany), MgSO4 (Merck, Germany), K2HPO4 (Merck, Germany); KH2PO4 (Merck, Germany), glycerol (Merck, Germany), Triton X-100 (Merck, Germany), imidazole (Sigma- Aldrich, USA), ampicillin (Biobasic, USA), kanamycin (Biobasic, USA), Isopropyl β-D-1-thiogalactopyranoside (IPTG, Biorad, USA), acrylamide (Sigma-Aldrich, USA), bis-acrylamide (Sigma-Aldrich, USA), Tris-HCl (Merck, Germany), tris base (Merck, Germany), HCl (Merck, Germany), Sodium Dodecyl Sulfate (SDS, Bio-Rad, USA), Ammonium persulfate (APS, Bio-Rad, USA), N, N, N’, N’- tetramethylethylenediamine (TEMED, Bio-Rad, USA), Tris (Bio- Rad, USA), 2-Mercaptoethanol (Bio-Rad, USA), Bromophenol Blue (Bio-Rad, USA), Glycine (Bio-Rad, USA), Coomassie Brilliant Blue R-250 (Bio-Rad, USA), methanol (Sigma-Aldrich, USA), glacial acetic acid (Merck, Germany), PageRuler™ Prestained Protein Ladder (10- 170 kDa, Thermoscientific, USA), BamHI and HindIII restriction enzymes (Promega, USA), BamHI_PirA_1F and HindIII_PirA_336R primers (Phusa Biochem, Vietnam).
Cloning and sequencing of PirA gene
The PirA gene was isolated from Vibrio parahaemolyticus strain K5 causing AHPND on white leg shrimp by PCR method with specific primers. A shotgun sequencing of bacterial small subunit ribosomal DNA gene fragments amplified from AHPND-infected shrimp revealed that the bacterial sequences of VpAHPND strains were not related to sequences normally found in diseased shrimp infected with other Vibrio bacterial species. Phylogenetic analysis showed that the strains were genetically diverse and not derived from a single genetic lineage. On the other hand, a preliminary sequence assembly and comparison analysis of the strains suggested that the target sequences originated from plasmid. The identified contigs of the strains were found to be homologous not to chromosomes of known V. parahaemolyticus but to the contigs obtained from other VpAHPND strains. Furthermore, a contig encodes the homologues of type IV pilus protein and conjugal transfer protein in the strain was also revealed, which suggests that it is located on a plasmid to insert the PirA gene into the pQE30 expression vector, specific primers BamHI and HindIII restriction enzymes were added (BamHI_PirA_1F forward primer: 5’-GGATCCATGAGTAACAATATAAAACATG-3'; HindIII_PirA_336R reverse primer: 5’-AAGCTTAGTGGTAATAGATTGTACAG-3').
PCR was carried out in 12 µl reactions containing 1 µl of genomic DNA template, 1 µl of each primer (10 pmol), 6 µl of 2X GoTaq® Green Master Mix (Promega, USA) and 4 µl of nuclease-free water for the amplication of the pirA gene. The reactions were run on MJ MiniTM Personal Thermal Cycler (BioRad, USA). PirA gene was amplified using the following parameters: Initial denaturation at 95°C for 5 minutes, 30 cycles of 95°C for 30 seconds, 53°C for 30 seconds and 72°C for 1 minute 30 seconds, followed by a final extension at 72°C for 10 minutes and store at 4°C. The PCR product was electrophoresed on a 1% agarose gel, voltage of 80 V in 1X TAE buffer with SafeView dye (1X TAE: SafeView=20:1). Electrophoresis result was observed on Ultra Slim LED Illuminator (Miulab, China).
The PCR product was cloned in pGEM®-T easy vector and transformed into E.coli TOP10 host cells by heat shock method [22]. The E. coli were grown for 24 to 36 h on LB (NaCl 10 g/L, Yeast extract 5 g/L, Peptone 10 g/L) agar medium supplemented with 100 µg/ml ampicillin+100 mM IPTG+200 mg/ml X-Gal (LB agar/ampicillin/IPTG/X-Gal medium) and the best grown were selected. Five white colonies were randomly selected and PCR with BamHI_PirA_1F and HindIII_ PirA_336R primers to screen E. coli cells containing pGEM-T easy/ PirA recombinant plasmid was performed. The recombinant plasmid was isolated from E. coli cells using EZ-10 Spin Column Plasmid DNA Mini-preps Kit (Biobasic, USA). PirA gene from pGEM-T easy/PirA plasmid were sequenced by Sanger’s method with specific primers (T7 promoter: 5'-TAATACGACTCACTATAGGG-3' and SP6Long: 5'- ATTTAGGTGACACTATAGAATAC-3') (Firstbase, Malaysia). The pirA gene sequencing was performed by First TBASE Laboratories Sdn Bhd (Selangor, Malaysia) after cloning. The sequences of PirAgene were analyzed by BioEdit software (ver 7.0.5.3) and compared with PirA gene sequences published on GenBank (accession number: KU556825.1) using the BLAST program.
Insertion PirA gene into expression vector
Recombinant pGEM-T easy/PirA plasmid and pQE30 expression vector were extracted from E. coli TOP10 cells and digested by BamHI and HindIII restriction enzymes (incubated at 37°C for 4 hours). Subsequently, the digest products were electrophoresed on 2% agarose gel; pQE30 vector and PirA gene were collected from gel and purified by ISOLATE II PCR and Gel Kit (Bioline, UK). The two purified products were ligated by T4 DNA ligase (Biobasic, USA) according to the molar ratio between pQE30 vector and PirA gene, which is 1:3, at 22°C for 1 hour to form a pQE30/PirArecombinant plasmid. Then the pQE30/PirAwas transformed into E. coli TOP10 cells using the heat shock method before selection on LB agar medium supplemented with 100 µg/ml ampicillin. We screened E. coli TOP10 cells containing pQE30/PirA recombinant plasmid by colony PCR method with BamHI_PirA_1F and HindIII_PirA_336R primers, as described in the previous section (See Cloning and Sequencing of pirA gene).
Creation of E. coli M15 cells containing recombinant pQE30/ PirA plasmid
Recombinant pQE30/PirA plasmid was extracted from E. coli TOP10 cells by EZ-10 Spin Column Plasmid DNA Mini-preps Kit (Biobasic, USA) and transformed into E. coli strain M15 cells by heat shock method [22]. Five transformed colonies grown on LB agar supplemented with 100 g/ml ampicillin and 50 g/ml kanamycin were randomly selected and further cultured in liquid LB medium supplemented with 100 g/ml ampicillin and 50 µg/ml ampicillin for recombinant plasmid extraction. Recombinant pQE30/PirA plasmids were extracted and checked for the presence of the PirA gene by PCR with BamHI_PirA_1F and HindIII_PirA_336R primers, as described previously. E. coli M15 cells carrying recombinant pQE30/PirAplasmid were selected for PirA gene expression.
Expression of recombinant PirA protein
E. coli M15 cells containing recombinant expression vector were cultured in 5 ml of LB medium supplemented with 100 mg/L ampicillin and 50 mg/L kanamycin, shaken at 180 rpm overnight at 37°C. Culture solution was transferred into 250 mL erlenmeyer flasks containing 100 ml of LB/ampicillin/kanamicin medium (4% v/v), incubated at 37°C with shaking speed of 200 rpm. For optimization of recombinant PirA antigen expression, cultivations were performed under different IPTG (isopropyl-β-D-hiogalactopyranoside) concentrations. In order to optimize IPTG concentration for recombinant PirA antigen expression in E. coli M15, different IPTG concentrations (0.1-1.0 mM) were added and cultured after reaching the desired OD600 nm of 1.0 at 37°C, 200 rpm. The samples were collected after 4 hours of the induction. Cell density was determined by measuring the optical absorbance of a sample at 600 nm (OD600 nm) on the double beam spectrophotometer U-2900 (HITACHI, Japan).
Extraction of recombinant antigens and Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
The recombinant E. coli M15 cell biomass was collected by centrifugation at 12,000 rpm for 2 minutes and then re-suspended in a Lysis buffer (50 mM potassium phosphate (pH=7.8), 400 mM NaCl, 100 mM KCl, 10% glycerol, 0.5% Triton X-100, 10 mM imidazole). The cell was homogenized by ultrasonic cell disruptor with 05 cycles (60 seconds of ultrasound and 30 seconds of rest), on sonicator at a frequency of 20 khz. Next, the sample was centrifuged at 12,000 rpm for 10 minutes to separate soluble and insoluble proteins. The supernatant was collected and assayed on 15% SDS-PAGE gel. The gel was stained with Coomassie Brilliant Blue for 30 minutes. Finally, the gel was washed with a washing solution (30% (v/v) methanol, 10% (v/v) acetic acid) until the gel became clear and protein bands appear (blue).
The recombinant E. coli M15 colony with high PirA expression was collected and used for the next experiments. The cells were cultured in the above conditions for the recombinant PirA gene expresstion. Recombinant PirA solution as purified by HisTrap FF column (GE Healthcare, USA) according to the manufacturer's instructions. The purified protein concentration was determined by the Bradford method [23].
Data analysis
Data were statistically analyzed using Minitab software version 16.2.0 and Microsoft Excel 2013 to calculate the mean and standard deviation. ANOVA was used to identify significantly different means compared between elution fractions with Tukey’s test at a probability level of P ≤ 0.05.
Sequencing of PirA gene
The amplification product of the PirA gene from Vibrio parahaemolyticus strain K5 was cloned into the pGEM-T easy vector (Promega, USA). We screened E. coli TOP10 colonies containing pGEM T easy/the PirA recombinant plasmid on LB agar/ ampicillin/IPTG/X-Gal medium. To confirm the presence of PirA gene in E. coli TOP10 colonies, five white colonies were randomly selected to conduct colony PCR with BamHI_PirA_1F and HindIII_PirA_336R primers. The PCR product electrophoresis showed a band of about 347 bp, matching the size of the PirA gene, and two nucleotide sequences of BamHI and HindIII restriction enzymes according to theoretical calculations in the protocol. Next, two E. coli TOP10 colonies containing pGEM T easy/PirA recombinant plasmid were selected for extraction the recombinant plasmid that provided material for the nucleotide sequence analysis of PirA gene. The results of PirA gene sequencing are shown in Figure 1. The analysis results showed that PirA gene has a length of 336 bp (Figure 1) (excluding two recognizable nucleotide sequences of BamHI enzyme at the 5 'end and the HindIII enzyme at the 3' end) and PirA gene sequence from Vibrio parahaemolyticus strain K5 is 100% similar to PirA gene from Vibrio parahaemolyticus strain V.03 which has been published on GenBank (accession number: KU556825.1).
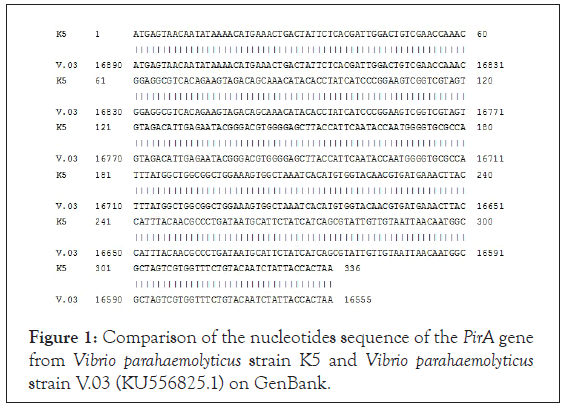
Figure 1: Comparison of the nucleotides sequence of the PirA gene from Vibrio parahaemolyticus strain K5 and Vibrio parahaemolyticus strain V.03 (KU556825.1) on GenBank.
Creation of pQE30/PirArecombinant plasmid
The PirA gene and after the pQE30 vector were collected and purified from digestion products by BamHI and HindIII restriction enzymes, and after the pQE30 vector and PirA gene were paired by the enzyme T4 DNA ligase to create the pQE30/ the PirA recombinant plasmid. The coupling product was then transformed into E. coli strain TOP10 and first screened on LB Agar medium supplemented with 100 ug/ml ampicillin. The transformation result showed that white colonies appeared on LB agar/ampicillin medium (Figure 2A). This indicates a successful transformation, and these colonies may contain pQE30/PirA recombinant plasmid. We randomly selected five colonies for biomass culture and plasmid extraction. PCR was performed with the extracted DNA plasmid template, and BamHI_PirA_1F and HindIII_PirA_336R primers to confirm the presence of the PirA gene. The PCR product was electrophoresed on 1% agarose gel (Figure 2B). The results of the study showed that PCR products from five selected colonies all appeared as bands of about 350 bp, matching the size of the PirA gene (336 bp) the addition of the recognizable nucleotide sequences of BamHI enzyme at the 5 end, and HindIII enzyme at the 3' end.
The PirA recombinant antigen expression in E. coli
The pQE30 expression vector carrying PirA gene was transformed into E. coli M15 cells by heat shock method. The transformed cells were selected randomly to extract recombinant plasmid and to check for the presence of the target gene (PirA) by PCR with BamHI_PirA_1F and HindIII_PirA_336R primers. The PCR products were electrophoresed on 1% agarose gel and showed the appearance of DNA bands with the size of about 347 bp, matching the size of PirA gene with addition of the recognizable nucleotide sequences of BamHI and HindIII enzymes (Figure 2B) The results showed that pQE30/PirA recombinant plasmid was successfully transformed into E. coli M15 cells. The cells carrying pQE30/PirA recombinant plasmid were cultured and induced PirA antigen expression by IPTG. The SDS-PAGE electrophoresis results (Figure 3) showed that the protein fraction obtained in the experimental group was about 15 kDa, and this protein fractxion did not appear in the control group E. coli M15.
Figure 2: Results of transformation of pQE30/PirA recombinant plasmid into E. coli strain M15 cells. (A) E. coli M15 colonies-results of transformation of pQE30/PirA recombinant plasmid; (B) Electrophoresis of PCR products of PirA gene from 05 E. coli M15 colonies-results of transformation of pQE30/PirA recombinant plasmid. Cells did not carry pQE30/PirA recombinant plasmid.
Figure 3: Results of SDS-PAGE electrophoresis of PirA recombinant antigen expression in E. coli M15 cells. M: PageRulerTM Prestained Protein Ladder (10-170 kDa, Thermoscientific); O: E. coli M15 cells did not carry pQE30/PirA recombinant plasmid; PirA: E. coli M15 cells carry pQE30/PirA recombinant plasmid.
PirA recombinant antigen purification
To confirm the presence of PirA antigens fused with the 6xHis tag, total protein of E. coli M15 recombinant cells was used as material for the purification of the PirA-6xHis fusion protein by affinity chromatography method with HisTrap FF column (GE Healthcare, USA). The results of electrophoresis on the 15% SDS-PAGE gel (Figure 4) showed that 6xHis-PirA fusion protein was recovered in a purified form, with the size of about 15 kDa matching the theoretical size. Thus, the study result confirmed that the PirA gene was successfully expressed in the 6xHis-PirA fusion protein form. The SDS PAGE electrophoresis result (Figure 4) for the total protein extracted from E. coli M15 recombinant cells and 6xHis-PirAtarget protein in elution fractions from the HisTrap FF column (EF1-EF6) with Elution buffer (20 mM sodium phosphate, 0.5 M NaCl, 0.5 M imidazole, pH 7.4) showed that the majority of 6xHis-PirA fusion protein was collected in elution fractions from EF2 to EF6, while most of the basic proteins of E. coli M15 that was unable to bind to Ni2+ ions on the HisTrap FF column were removed during wash step with the binding buffer (20 mM sodium phosphate, 0.5 M NaCl, 5 mM imidazole) previously. Most of the protein 6xHis-PirA binding on the column was collected from EF2 to EF5 elution fractions, while 6xHis-PirA protein was collected very little at EF1 and EF6 elution fractions. Thus, the 6xHis-PirA protein has been purified successfully. However, there is still a small amount of impurity protein in the purified product in EF1 and EF2 elution fractions.
Figure 4: Electrophoresis of 6xHis-PirA fusion protein after purification with HisTrap FF column. M: PageRulerTM Prestained Protein Ladder (10-170 kDa, Thermoscientific); O: The total protein of E. coli M15 recombinant cells; EF1 to EF5: 6xHis-PirA fused protein in elution fractions (1-5) from HisTrap FF column.
Analytical results by the Bradford method (Table 1) showed that the concentration of 6xHis-PirA protein in elution fractions from the HisTrap FF column was different. In which, EF3 elution fraction collected the highest target protein concentration (1,586.54 µg/ ml), followed by EF2 elution fraction (1,317.1 µg/ml). The collected protein concentrations decreased gradually from EF3 to EF6 elution fractions, reaching the lowest in EF6 (17.46 µg/ml).
| Elution fractions | EF1 | EF2 | EF3 | EF4 | EF5 | EF6 |
|---|---|---|---|---|---|---|
| 6xHis-PirA fusion protein concentration | 67.92 ± 1.21a | 1,317.14 ± 6.27b | 1,586.54 ± 5.53a | 1,001.09 ± 2.17c | 354.04 ± 0.60d | 17.46 ± 0.60f |
Note: The means with different letters a,b,c,d,f within the same row are significantly different between elution fractions at the 0.05 probability level.
Table 1: 6xHis-PirA fusion protein concentration was collected in elution fractions from HisTrap FF column (μg/ml).
The PirA gene sequence in this study is like to those of analysis of V. parahaemolyticus strains causing AHPND isolated in Baja California Sur and Sinaloa (Mexico). That study showed that these strains carry the PirAvp gene with the size of 336 bp [24]. Results of genomic analysis of V. parahaemolyticus strain MSR16 and V. parahaemolyticus strain MSR17 isolated from Penaeus monodon in southwestern Bangladesh showed that both strains carry the plasmid (~69 Kbp), which contained PirA and PirB toxin genes. The PirA gene's size was 336 bp (starting at position 64,962 bp and ending at position 65,297 bp on the plasmid of MSR16 strain; Starting at position 63,108 bp and ending at position 63,443 bp on the plasmid of MSR17 strain) [21]. The results of genomic analysis of V. parahaemolyticus strain 13-028/A3 showed that the toxin proteins were encoded by two 2 genes (pirA-like and pirB-like) in a segment 3.5 kb, the pirA-like gene (336 bp) and the pirB-like gene (1,317 bp) encoded for 13 kDa and 50 kDa proteins, respectively [17].
There are several protein expression systems for production of recombinant proteins such as bacterial, yeast, insect, mammalian systems. In this study, E. coli strain M15 was used as a host cell to express PirA antigen of Vibrio parahaemolyticus strain K5 because of E. coli’s outstanding properties including easy manipulation, inexpensive culture medium and rapid growth [25-32], high performance of recombinant protein expression [26], and easy collection target proteins [33]. In addition, antigens synthesized by E. coli host cells are suitable materials for antibody production [34]. Numerous studies have been performed to optimize the expression conditions of recombinant proteins in E. coli [32,35-39].
Our research results showed that the expression of PirA gene was produced in the form of a fusion protein with high concentration. The molecular weight of this fusion protein was approximately 15 kDa (including 6xHis tag of pQE30 vector) (Figure 3). This size is higher than found in study by Han JE, et al. [17], where proteomic analysis showed that the pirA-like genes (336 bp) encode for a protein with the size of 13 kDa. Lee CT, et al. [18], performed western blot analysis with antibodies against PirAvp from AHPND causing V. parahaemolyticus and found that this strain secreted PirAvp (12 kDa) into the culture medium after 1 h after cultivation. The variation in the size of the PirA protein may be due to the influence of several factors during SDS PAGE electrophoresis and the effect of the expression system on the PirA gene expression. The PirA antigen was expressed by pQE30 expressing vector system and was fused with six amino acids histidine. This 6xHis tag has a high affinity for Ni2+ ions on the Histrap FF column (GE Healthcare, USA), allowing for easy collection of 6xHis-PirA fusion protein and removal of other proteins of E. coli M15 cell when their total proteins are passed through the chromatographic column. The 6xHis-PirA fusion protein recovered from the Histrap FF column had high purity and concentration (Figure 4 and Table 1), and can be used as a raw material for further research to produce antibodies to prevent AHPND in shrimp caused by Vibrio spp.
The study results showed that PirA gene of Vibrio parahaemolyticus strain K5 was successfully cloned in pGEM-T easy vector. The PirA gene is 336 bp in size and 100% similar to the nucleotide sequence of PirA gene of Vibrio parahaemolyticus strain V.03 that has been published on GenBank (KU556825.1). The PirA gene was successfully inserted into a pQE30 expression vector. The PirA recombinant antigen was expressed as a 6xHis-PirA fusion protein (about 15kDa) in E. coli strain M15 and the purified protein was obtained at high concentrations by affinity chromatography method with HisTrap FF column. The purified PirA recombinant protein can be used as raw material for further research to develop antibodies to prevent acute hepatopancreatic necrosis disease in shrimp.
https://csdlkhoahoc.hueuni.edu.vn/index.php/topic/index/user/109/page/1, license of Ministry of Education and Training, Vietnam. Cloning and expression of PirA gene of Vibrio parahaemolyticus strain K5 causing acute hepatopancreatic necrosis disease in whiteleg shrimp in E. coli host cell. Genbank: https://www.ncbi.nlm.nih.gov/nuccore/MN846072.1/, https://www. ncbi.nlm.nih.gov/nuccore/MN846073.1/
-1st_BASE_3641594_A4(K5) T7promoter.ab1: The nucleotide peaks of pirA gene form T7 promoter primer from Vibrio parahaemolyticus strain K5 causing AHPND in whiteleg shrimp in Phong Dien district, Thua Thien Hue province, Vietnam.
-1st_BASE_3641594_A4(K5)_T7promoter.seq: The nucleotide sequence of pirA gene form T7 promoter primer from Vibrio parahaemolyticus strain K5 causing AHPND in whiteleg shrimp in Phong Dien district, Thua Thien Hue province, Vietnam.
-1st_BASE_3641598_A4(K5)_SP6Long.ab1: The nucleotide peaks of pirA gene form SP6Long primer from Vibrio parahaemolyticus strain K5 causing AHPND in whiteleg shrimp in Phong Dien district, Thua Thien Hue province, Vietnam.
-1st_BASE_3641598_A4(K5)_SP6Long.seq: The nucleotide sequence of pirA gene form SP6Long primer from Vibrio parahaemolyticus strain K5 causing AHPND in whiteleg shrimp in Phong Dien district, Thua Thien Hue province, Vietnam.
-6xHis-PirA_fusion_protein_concentration.xlsx: Concentrations of 6xHis-PirA fusion protein extracted from E. coli M15 recombinant cells.
This investigation was funded by the Ministry of Education and Training, Vietnam, through a ministry-level science and technology project (Reference: CT2018-DHH-04).
The authors declare no conflicts of interest.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: van Khanh N, Dung TQ, Khanh van TQ, Linh NQ (2022) Cloning and Expression of PirA Gene of Vibrio parahaemolyticus Strain K5 Causing Acute Hepatopancreatic Necrosis Disease in Whiteleg Shrimp in E. coli Host Cell. J Clin Toxicol. 12:522.
Received: 30-Sep-2022, Manuscript No. JCT-22-19422; Editor assigned: 03-Oct-2022, Pre QC No. JCT-22-19422 (PQ); Reviewed: 17-Oct-2022, QC No. JCT-22-19422; Revised: 24-Oct-2022, Manuscript No. JCT-22-19422 (R); Published: 31-Oct-2022 , DOI: 10.35248/2161-0495.22.12.522
Copyright: © 2022 van Khanh N, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.