Research Article - (2020)Volume 5, Issue 1
Although MAIT cells regulate the pathogenesis of various inflammatory diseases, their roles in the development of colorectal cancer are still unclear. The objective of this study is to investigate the level of circulating MAIT cells and the level of expression of the membranous KIRs receptors among CRC patients and control subjects. A total number of 89 subjects were recruited, 46 of them represented the patients who were diagnosed with colorectal cancer during the study. The remaining 43 subjects represented normal healthy control. The cases in this group were blood bank donors who visited the same medical center and did not have a known personal or family history of cancer. Peripheral blood mononuclear cells (PBMCs) were isolate and of MAIT cells were phenotypically identified by flow cytometry using various monoclonal antibodies. The presence of HLA-C1 and HLA-C2 groups were typed by PCR.
High frequency of HLA-C2 among patients with CRC (87%) compared with control subjects (74.4%). The frequency of HLA-C1 was higher in control subjects (72.1%) compared with CRC patients (65.2%). In addition, genotype distribution showed that the frequencies of HLA-C1C2 washigher in CRC patients (52.2%) compared with control subjects (48.8%). Although statistically insignificant, the percentage of MAIT cells were higher in CRC patients compared to control subjects. The percentage of MAIT cells was higher in patients with CRC in stage III and IV, but lower in stage II compared with control subjects. The protein expression of MAIT cells associated phenotyping antigens and some KIR receptors such as CD45RA, CD45RO, CD62L, CD11a, CD158a, CD158b, CD158e and CD158f were reported. A relatively lower percentage of CD45RA expression was seen in CRC patients compared with control subjects. There was a significant reduction in CD45RO, CD62L, CD158a, CD158e and CD158f expression in CRC patients. Stratification analysis of 46 CRC patients indicated that the percentages of circulating MAIT cells were lower in stage II and higher in stages III, and IV. The frequencies of circulating MAIT cells had not been reduced in patients with colorectal cancer.
Colorectal cancer; MAIT cells; KIR gene
Colorectal cancer (CRC) is a major contributor to cancer-related morbidity and mortality. About one million new CRC cases occur annually with more than half a million deaths worldwide [1]. Despite a relatively low incidence, CRC is ranked the second most common type of cancer in Saudi Arabia among males (10.6%) and the third among females (8.9%) [2]. Currently, the main therapeutic approaches for treatment are surgery, chemotherapy, and radiotherapy [3]. The most recent progress in immunology and biotechnology haveopened new perspectives for the development and implementation of immunotherapy preventing cancer and infectious diseases [4]. Still, despite of this progress, there are a number of vague parts of this cancer that require further research. As such, researchers are expected to identify the markers, in which anti-tumor impact can be predicted and used to survive the patient before immunotherapy.
Mucosal Associated Invariant T (MAIT) cells are a unique innate-like lymphocyte cell subset that express a semi-invariant TCR Vα7.2-Jα33 chain and can be identified in human beings by expressing Vα 7.2 segment together with CD161 surface molecule or IL-18Rα [5-7]. In humans, MAIT cells are located in the intestinal mucosa and liver in preference to other tissues [8-10]. Unlike traditional MHC-1, limited to T cell system, MAIT cells can recognize and respond to vitamin B2 (riboflavin) biosynthesis precursor derivative presented by major histocompatibility complex (MHC) related protein 1 (MR1) [11-13]. Upon activation in the presence of bacterium-infected APC, MAIT cells release various cytokines, including IFN-γ, TNF-α and IL-17A [7,14,15].
MAIT cells are important for immune defense against infection by pathogens and regulate the pathogenesis of various inflammatory diseases [16]. Although, there are many studies in relation to the impact of MAIT cells on pathogenic infections and autoimmune diseases, only a few researches have been conducted to study the impact of these cells on the development of colorectal cancer (CRC), which is not defined clearly. Thus, it has been reported that the analysis of Killer-cell immunoglobulin-like receptor (KIR) gene content is correlated with the particular cancer [17].
Several studies have described KIR/HLA compound genotypes that associate with susceptibilities to certain cancers [18]. Higher levels of KIR-mediated inhibition may also facilitate tumor escape, as has been shown for melanoma where the compound genotype KIR2DL2/2DL3/HLA-C1 was more frequent in patients as compared to controls [19].
It has been found that, the entire inhibitory genes combined with their ligands were more frequent in colorectal cancer patients than in controls, and there was an association in KIR2DL3 and HLA-C1 and colorectal cancer patients. The development of colorectal cancer and the strength of this association increases when KIR2DL3 and KIR2DL2 are accumulated together with the C1 [20].
CD158a or (KIR2DL1) is one of several immunoglobulin-like receptors that bind MHC class I molecules and transmit inhibitory signals [21,22]. CD158b or KIR2DL2/3, expressed on natural killer cells and a subset of T cells, is a member of the KIR family with inhibitions role. CD158e2, or expresses on the surface of NK cells among approximately 38% of population, where HLA-Bw4 molecules serve as ligands for KIR3DS1 [23]. KIR3DS1 together with HLA-Bw4 alleles are associated with slower disease progression [24]. CD158f or KIR2DL5A clearly inhibitory potency and clonal expression which occur on NK cells and T-lymphocytes [25].
The objectives of this study is to investigate the connection between the level of circulating MAIT cells and the level of expression of the membranous KIRs receptors by using flow cytometry and (SSP-PCR) among CRC patients and control subjects.
Subjects
This is a prospective case-control study, conducted in King Fahad Specialist Hospital-Dammam (KFSHD), in the Eastern Province of Saudi Arabia. The appropriate ethical approval was obtained from the local research ethics committee under Institutional Review Board Number (EXT0328) during 2016-2018. A total number of 89 subjects were recruited, 46 of them represented the patients who were diagnosed with colorectal cancer during the study. A consultant oncologist at KFSHD oncology center completed staging and detailed therapy plan among these 43 patients. The remaining 43 subjects represented normal healthy control. The cases in this group were blood bank donors who visited the same medical center and did not have a known personal or family history of cancer. All subjects were informed and consented before proceeding with the study.
Isolation of peripheral blood mononuclear cells (PBMCs) and the identification of MAIT cells
Peripheral blood samples were collected from each colorectal cancer subject and healthy control using a standard venipuncture procedure and collecting in heparin-containing tubes. PBMC were isolated by Ficoll-paque Plus (GE Healthcare) and density-gradient centrifugation. MAIT cells were identified phenotypically as CD3+, CD161+/TCR7.2+, TCR γδ- [6,26].
Monoclonal antibodies (mAbs) and flow cytometry
The monoclonal antibodies used were CD3-PerCP (Cat No: 130-094-965), CD161-peVio 770 (Cat No:130-099-965), Anti- TCR Vα7.2-APC-Vio700 (Cat No:130-100-179), Anti-TCRγ/ δ-APC, human, (Cat No:130-096-866), CD45RA-PE, human, (Cat No:130-098-184), CD45RO-PE, human, (Cat No: 130-109-508), CD62L-PE, human, (Cat No:130-099-717), CD11a-PE, human, (Cat No:130-105-479), CD158a (KIR2DL1)- PE, human, (Cat No:130-103-967), CD158b-PE, human, (Cat No:130-099-397), D158e(KIR3DL1)-PE, human, (Cat No: 130-092-473), CD158f (KIR2DL5)-PE, human, (Cat No: 130-115-911). The antibodies were obtained from MiltenyiBiotec, BergischGladbach, Germany.
Data collection and analysis of flow cytometry
Immunophenotyping was performed on a FACS Canto II instrument (Becton Dickinson, San Jose, CA, USA). Collection and analysis were performed using FACSDiva v8.0 software. The forward scatter light (FSC) and the side scatter light (SSC) parameters were adjusted to each sample in order to observe all cell types. The cell collection rate was adjusted to 500-1000 events/second. Small particles and debris were excluded by FSC gate on lymphocytes. A fluorescence threshold was set at 5000 in order not to not exceed 0.5% of background fluorescence.
MAIT cell analysis was completed using a sequential gating strategy. Staring from gating on CD3 positive population to target T-lymphocytes, then T-cells expressing TCR gd were filtered by gating on the CD3+/TCR gd + population. Subsequently, the MAIT cells were gated as a population CD161 +/TCR7-2+, of which MAIT associated phenotyping antigens (i.e. CD45RA, CD45RO, CD62L, CD11a, CD158a, CD158b, CD158e and CD158f) were analyzed for percentage of expression and mean fluorescent intensity.
HLA-C1 and C2 Genotyping
For each reaction, 50-100 ng of DNA was used in a 15 μL final volume. For 16 KIR genes, HLA-C1 and HLA-C2 group typing, the same primers were used as reported before [19,27]. For each reaction, positive, negative, and internal controls were used. All PCR reactions were performed with the thermocycler apparatus T100TM (Thermal Cycler from Bio-Rad Laboratories, Inc. Life Science Research, California 94547, United States, and Veriti Thermal Cycler from Thermo Fisher Scientific, United States).
Statically analysis
Different types of tests were used for the phenotype and functional studies. For example, unpaired t-test and paired t-test were used when the data had assumed a Gaussian distribution. In contrast, for the data that did not assume a Gaussian distribution a Mann-Whitney U test was used. Groups with a p value of<0.05 were considered to be statistically significantly different. Statistical analysis was conducted using software Graph pad prism version 7 for the phenotyping and functional studies.
Samples were first classified according to their BMI based on height and weight correlation.
We have examined Saudi patients with CRC, of which 30 males and 16 females, and Saudi control subjects consisted of 27 males and 16 females. The mean ages of participants were 56.37 ± 12.93 years in males, and 55.69 ± 13.05 years in females. In both groups the height, weight and BMI did not show significant differences: in CRC patients, the mean height was 1.7 ± 0.08 in males and 1.55 ± 0.07 in females. Subsequently, the mean weight was 84.85 ± 20.13 in males and 81.94 ± 23.95 in females. BMI was 29.30 ± 5.95 and 34.04 ± 9.27, respectively. The mean height was 1.69 ± 0.05 in males and 1.57 ± 0.05 in females among control subjects’ group. The mean weight was 83.59 ± 11.73 in males and 75.44 ± 22.59 in females. The mean BMI was 27.92 ± 6.93 and 30.32 ± 7.29, respectively.
Distribution of the KIR ligand C1/C2 in patients and control groups
The distribution of HLA-C groups and genotypes are shown in (Table 1). Statistical analysis showed a high frequency of HLA - C2 among patients with CRC (87%) compared with control subjects (74.4%). The frequency of HLA-C1 was higher in control subjects (72.1%) compared with CRC patients (65.2%). In addition, genotype distribution showed that the frequencies of HLA-C1C2 was higher in CRC patients (52.2%) compared with control subjects (48.8%) (Table 2).
| Variables | Control Subjects | CRC Patients | ||
|---|---|---|---|---|
| Men | Women | Men | Women | |
| Number of Subjects | 27 | 16 | 30 | 16 |
| Age | 54.74 ± 11.55 | 54.94 ± 11.53 | 56.37 ± 12.93 | 55.69 ± 13.05 |
| Height | 1.69 ± 0.05 | 1.57 ± 0.05 | 1.70 ± 0.08 | 1.55 ± 0.07 |
| Weight | 83.59 ± 11.73 | 75.44 ± 22.59 | 84.85 ± 20.13 | 81.94 ± 23.95 |
| BMI | 27.92 ± 6.93 | 30.32 ± 7.29 | 29.30 ± 5.95 | 34.04 ± 9.27 |
Table 1: The characteristics of CRC patients and Control subjects of the study.
| Genotype | Patient | Control | OR | CI 95% | Chi square and P-value |
|---|---|---|---|---|---|
| C1C1 | 6(13.04) | 10(23.3) | 0.495 | 0.163-1.505 | Χ2=1.57; P-value=0.21 |
| C2C2 | 16(34.8) | 11(25.6) | 1.552 | 0.62–3.87 | Χ2=0.89; P-value=0.35 |
| C1C2 | 24(52.2) | 21(48.8) | 1.14 | 0.50–2.63 | Χ2=0.1; P-value=0.75 |
| C1 | 30(65.2) | 31(72.1) | 0.73 | 0.29–1.79 | Χ2=0.49; P-value=0.48 |
| C2 | 40(87) | 32(74.4) | 2.29 | 0.76–6.87 | Χ2=2.26; P-value=0.13 |
Table 2: The frequencies and percentage of the KIR ligand C1/C2 in CRC patients and control.
MAIT cells frequencies in blood of CRC patients and healthy subjects
The presence of MAIT cells in the peripheral blood of CRC patients and control subjects were evaluated by identifying the expression of CD3+, CD161+/TCR7.2+ on their surface. Four-color flow cytometry was used to determine the percentage of MAIT cells. The lymphocytes populations were gated on forward and side scatter (Figure 1).
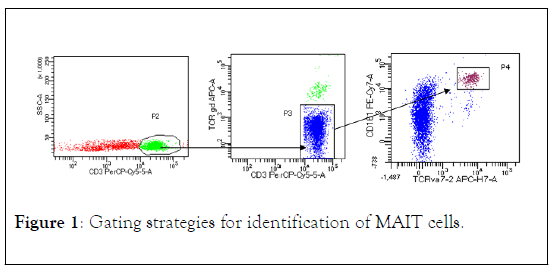
Figure 1: Gating strategies for identification of MAIT cells.
The percentage of MAIT cells were higher in CRC patients compared to control subjects. However, the difference between the two groups did not reach statistically significant differences (Figure 2). The median value was higher in CRC patients (3.265) compared to control subjects (2.6).
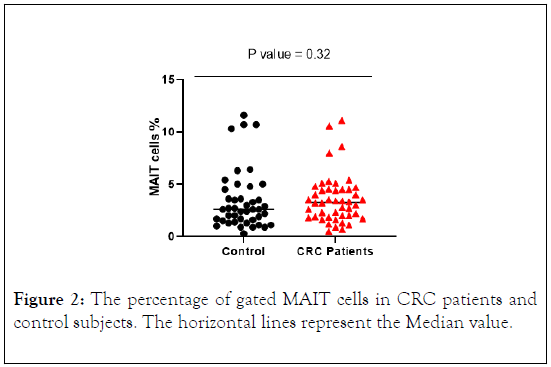
Figure 2: The percentage of gated MAIT cells in CRC patients and control subjects. The horizontal lines represent the Median value.
MAIT cells were stratified in patients with CRC by disease stage. The result showed that the percentage of MAIT cells was higher in patients with CRC in stage III and IV, but lower in stage II compared with control subjects (Figure 3). However, the difference between the four groups did not reach the statistical significant differences.
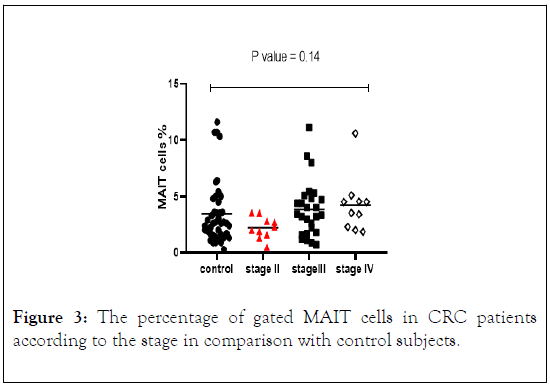
Figure 3: The percentage of gated MAIT cells in CRC patients according to the stage in comparison with control subjects.
MAIT cells phenotype antigen and KIR expression
The protein expression of MAIT cells associated phenotyping antigens and some KIR receptors such as (CD45RA, CD45RO, CD62L, CD11a, CD158a, CD158b, CD158e and CD158f) were analyzed for percentage of expression. A relatively lower percentage of CD45RA expression was seen in CRC patients compared with control subjects (Figure 4). There was a significant reduction in CD45RO expression in CRC patients in comparison with control subjects. CD11a expression indicated similarity in both patients and control with no statistically significant differences. However, CD62L analysis indicated a lower expression on MAIT cells in CRC patients compared with control subjects with significant difference (Figure 4). In CRC patients CD158a, CD158e and CD158f were expressed at lower percentage on MAIT cells in CRC patients and higher percentage was observed in control subjects with significant difference (p value = 0.01, 0.01, 0.03), also similar result was obtained for CD158b, however the results did not reach statistically significant differences (Figure 5).
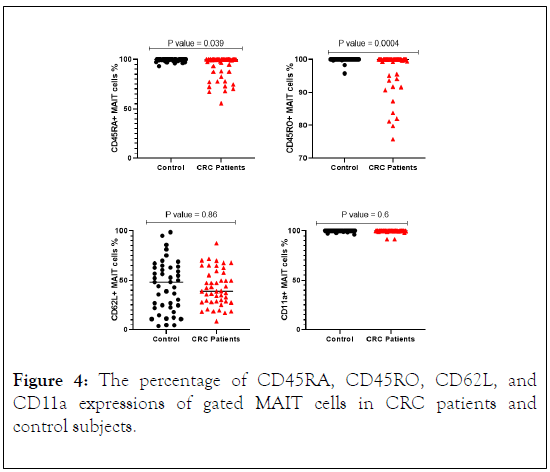
Figure 4: The percentage of CD45RA, CD45RO, CD62L, and CD11a expressions of gated MAIT cells in CRC patients and control subjects.
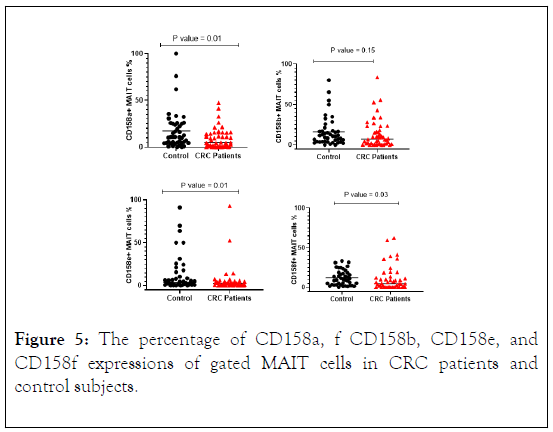
Figure 5: The percentage of CD158a, f CD158b, CD158e, and CD158f expressions of gated MAIT cells in CRC patients and control subjects.
Stratification analysis revealed that the percentage of MAIT cells that express CD158a, cd158e and CD158f were lower in CRC patients in all stages compared with control subjects. While CD158b was lower in CRC patients in stages III and IV and higher in stage II compared with control subjects. However, the difference between the four groups did not reach statistically significant differences (Figure 6).
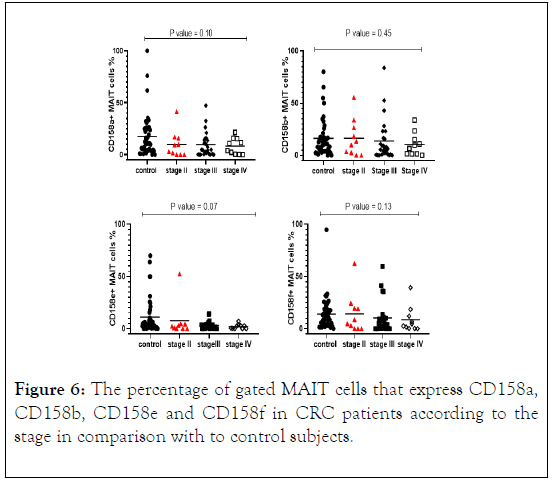
Figure 6: The percentage of gated MAIT cells that express CD158a, CD158b, CD158e and CD158f in CRC patients according to the stage in comparison with to control subjects.
KIR2DL2 and 2DL3 recognize HLA-C group 1 alleles, while HLA-C2 alleles are bound by KIR2DL1. Therefore, frequencies of KIR+ MAIT cells were measured in CRC patients and control subjects with or without the gene for the cognate ligand.
The percentage of MAIT cells expressing CD158a in the presence of HLA-C2 was lower in CRC patients compared with control subjects with significant differences (p value = 0.004). However, in the absence of HLA-C2, CD158a expression was higher in CRC patients (Figures 7a and 7b).
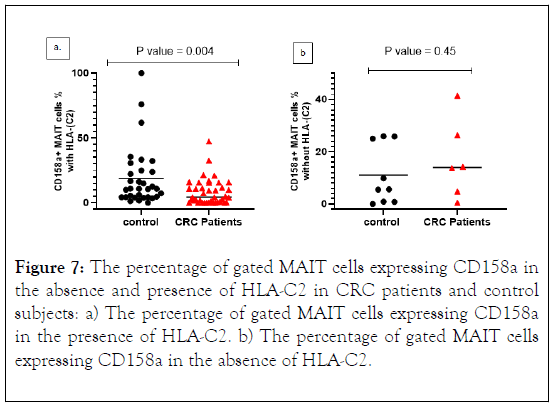
Figure 7: The percentage of gated MAIT cells expressing CD158a in the absence and presence of HLA-C2 in CRC patients and control subjects: a) The percentage of gated MAIT cells expressing CD158a in the presence of HLA-C2. b) The percentage of gated MAIT cells expressing CD158a in the absence of HLA-C2.
The percentage of MAIT cells expressing CD158b in the presence of HLA-C1 was lower in CRC patients compared with control subjects with no significant differences. In the absence of HLA-C1, CD158b expression was higher in CRC patients (Figures 8a and 8b).
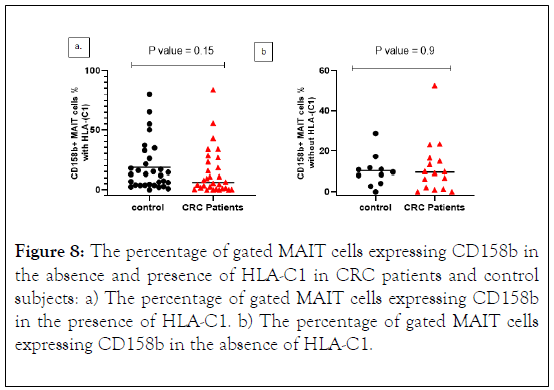
Figure 8: The percentage of gated MAIT cells expressing CD158b in the absence and presence of HLA-C1 in CRC patients and control subjects: a) The percentage of gated MAIT cells expressing CD158b in the presence of HLA-C1. b) The percentage of gated MAIT cells expressing CD158b in the absence of HLA-C1.
KIR2DL2 and 2DL3 recognize HLA-C group 1 alleles while HLA-C2 alleles are bound by KIR2DL1. Therefore, frequencies of KIR+ MAIT cells were measured in CRC patients with the gene for the cognate ligand according to the stage. The percentage of MAIT cells expressing CD158a in the presence of HLA-C2 was lower in CRC patients in stages II, III and IV compared with control subjects with statistically significant differences (p value=0.03) (Figure 9). Median levels of CD158a +MAIT cells indicate positive correlation with advancing cancer stages.
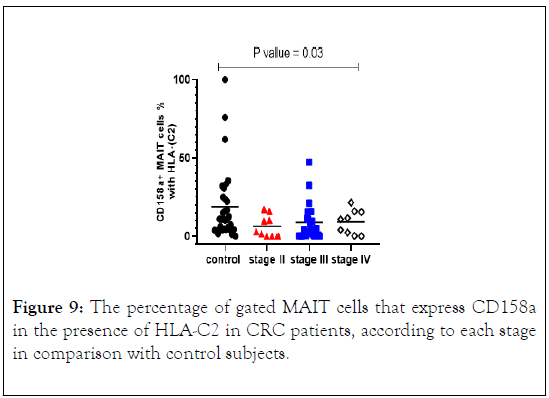
Figure 9: The percentage of gated MAIT cells that express CD158a in the presence of HLA-C2 in CRC patients, according to each stage in comparison with control subjects.
The percentage of MAIT cells expressing CD158b in the presence of HLA-C1 were quite similar in CRC patients in all stages and control subjects with no significant differences (Figure 10).
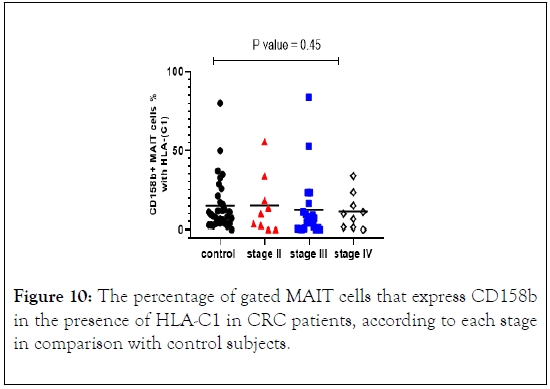
Figure 10: The percentage of gated MAIT cells that express CD158b in the presence of HLA-C1 in CRC patients, according to each stage in comparison with control subjects.
The number and level of circulating MAIT cells were evaluated in peripheral blood of CRC patients and control subjects. The percentage of MAIT cells in CRC patients was higher compared with control subjects.
Stratification analysis of 46 CRC patients indicated that the percentages of circulating MAIT cells were lower in stage II and higher in stages III, and IV. These results were consistent with a previous study, which showed that the frequencies of circulating MAIT cells had not been reduced in patients with different solid tumor [6]. However, current findings are in contradiction with alternative study that revealed a reduction in MAIT cells frequencies in patients with inflammatory bowel diseases (IBD) [28]. However, no statistically significant differences were found. Furthermore, the Median in stratification analysis revealed that the percentage of circulating MAIT cells increased with progression of the disease to advance stages, which did not correspond to what was previously demonstrated [29].
MAIT cells in CRC patients displayed a reduction in CD45RA, where naïve phenotype and results indicated a significant lower percentage. Furthermore, CD45RO expression was reduced again with a significant lower percentage. The memory phenotype CD45RO was expressed by circulating MAIT cells, while naïve phenotype CD45RA was negative [15,26], which was not consistent with our result for CD45RA. Further, the other study revealed that MAIT cells express CD45RA upon activation in patient with Tb [30].
Lymph node homing receptor CD62L expression was lower in CRC patients compared to control subjects, which was reflecting a memory phenotype with intermediate expression and no significant result. These findings are not consistent with similar studies, which indicated that MAIT cells egress the thymus into the blood, they recirculate between lymphatic system and acquire an effector memory phenotype, therefore, down regulate CD62L, and respond rapidly to re-stimulation, but do not express CD62L [15,31]. The expression of CD62L is required by T lymphocytes for entering a lymph node from the blood [32].
High expression of LFA-1 (CD11a) was shown in both CRC patients and control subjects with no statistically significant differences. These results are consistent with another study demonstrated overexpression of CD11a in patients with MS disease [33].
The frequency of functional KIR in MAIT cells was investigated in CRC patients and control subjects.
Anti-CD158a, b, e, and f antibodies were used to detect KIRs on the surface of MAIT cells in CRC patients and control subjects. The total percentage of CD158a+ MAIT cells was significantly lower in CRC patients compared with control subjects (P=0.01). Total CD158a+ MAIT cells and the median levels reduced with advancing stages. Stratification analysis of CD158a+ MAIT cells indicated a reduction with advancing cancer stages. The percentage of CD158a+ MAIT cells according to disease stage indicated a significantly reduction in stage III (P=0.04).
It has been reported that CD158a+NK cells frequency in healthy individuals who possessed its HLA-C2 ligand was higher than others lacking HLA-C2 alleles [34]. Furthermore, it was shown that NK cells expressed higher levels of CD158a and CD158b receptors in the presence of HLA-C. This was associated with susceptibility to tumors and disease progression [35,36]. Additionally, several studies revealed the involvement of KIR receptor and corresponding ligand in the pathogenesis, and clinical progression of various human diseases including cancer [37-39].
We have investigated whether there is a link between CD158a +MAIT cells proportion in the presence and absence of HLAC2.
A similar result for the frequency of CD158a+ MAIT cells with its ligand HLA-C2 was found in control subjects. In the absence of the ligand, the frequency of CD158a+ MAIT cells was higher in CRC patients and lower frequency in control subjects. Similar results for the median level with no statically significant results. However, the frequency significantly reduced in CRC patients (p value=0.004), with similar result for the median level.
We compared the frequency of CD158a+MAIT cells in all stages of CRC and control subjects in the presence of its ligand HLA-C2 and found a significantly reduction in all stages of CRC (p value=0.03) vs. control with gradually increase in the median, as the disease progress to advancing cancer stages.
Stratification analysis of CD158a+MAIT cells with its ligand HLA-C2 in CRC patients indicated that the frequency was lower in all stages with significant result in stage II (p value=0.02) and stage III (p value=0.01). The percentage of CD158b+ MAIT cells was lower in CRC patients compared with control subjects with no statically significant result.
We stratified CRC patients and found that CD158b+ MAIT cells were decreased as the diseases progress to advancing cancer stages. The median level was higher in stage II and gradually reduced in stage III and IV. CD158b+ MAIT cells in relation to its HLA-C1ligand expression shown a reduction in the frequency and median compared to control subjects. Whereas, in the absence of HLA-C1 the result revealed an increase in the frequency of CD158b+ MAIT cells in CRC patients vs. control subjects. The results of median level were analogous in both CRC patients and control subjects.
The percentage of CD158e+ MAIT cells were significantly lower in CRC patients compared to control subjects (p value=0.01) with a decreased in the median level with the progression of the disease. The median percentage of CD158f+ MAIT cells in CRC patients and control subjects were similar to statically significant results (p value=0.03) and reduced in CRC patients with the progression of the disease. Interestingly, when comparing the proportion of CD158a and b+ MAIT cells in all stages with control subjects, the results showed inverse correlation with cancer stages in CRC patients and the median gradually decrease in each stage.
The authors declare no conflict of interest.
Citation: Elsafi SH, Hassan MMA, Woodman A, Al Omar SY, Mansour L, Halawaani H, et al. (2020) Characterization of Circulating Mucosal Associated Invariant T cells in Colorectal Cancer Patients. Immunogenet Open Access. 5:126.
Received: 04-May-2020 Accepted: 18-May-2020 Published: 25-May-2020 , DOI: 10.35248/igoa.20.5.126
Copyright: © 2020 Elsafi SH, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.