Chemotherapy: Open Access
Open Access
ISSN: 2167-7700
ISSN: 2167-7700
Research Article - (2022)Volume 10, Issue 4
Background: It has been revealed that aging plays crucial roles in tumorigenesis, prognosis, and therapy response of tumors including esophageal carcinoma (ESCA). In this work, we aim to establish an aging-relevant risk signature to assess the survival outcome and immunogenicity status of ESCA patients.
Methods: A total of 351 ESCA patients with both gene expression data and clinical information from 3 independent datasets were curated. The Lasso-Cox regression model was applied to identify the aging genes that contributed most to the survival outcome. The risk signature was constructed by combining the specific gene expression level with the corresponding regression coefficients. Microenvironment-based immunologic factors, mutational burden, and significantly mutated genes were evaluated based on the identified risk groups. One cohort under the immune checkpoint inhibitor (ICI) treatment was used to investigate the immunotherapy predictive roles of the determined aging signature.
Results: Based on the 22 aging-relevant genes, a risk signature was constructed. ESCA patients with low-risk scores had improved survival outcomes in both discovery and validation datasets. Subsequent immunologic exploration demonstrated that the enhanced infiltration abundance of immune-response cells, decreased abundance of immune- suppressive cells, immune response-related signals, and the preferable ICI indicator enrichment were found in the low-risk group. Genomic mutation analysis showed the elevated mutational burden and increased mutation rates of significantly mutated genes of TP53, NAV3, and FAT1 were observed in patients with low-risk scores. In the ICI- treated cohort, we noticed that low-risk aging scores were significantly linked to favorable treatment outcomes and elevated response rates.
Conclusion: In summary, our identified aging risk signature showed the associations with survival, immune microenvironment, immunogenicity, and especially, the immunotherapy efficacy, which offers the clues for guiding prognosis evaluation and immune treatment strategies, and promotes precision therapy of ESCA patients.
Esophageal carcinoma; Aging risk signature; Survival outcome; Immune microenvironment; ICI response
Esophageal Cancer (ESCA) is the sixth leading cause of death across all cancers, accounting for 5% of all cancer-relevant deaths in 2018 [1]. ESCA is classified into esophageal squamous cell cancer and adenocarcinoma. Approximately half of ESCA patients present with unresectable or metastatic settings after diagnosing [2]. Over the past few years, owing to the improvements of multidisciplinary treatment, better therapy efficacies and clinical benefits for ESCA patients are observed. However, the survival outcome of unresectable or metastatic ESCA was still poor, with a median survival of less than one year [3]. Recent multiple molecular indicators were identified to predict ESCA prognosis outcomes; nevertheless, they are sometimes ineffective [4]. Therefore, more robust biomarkers are necessary to reliably evaluate the survival status of ESCA patients.
The invention of Immune Checkpoint Inhibitor (ICI) treatment has greatly improved the prognosis of several advanced cancers, such as melanoma, renal cancer, NSCLC, and ESCA [5-7]. Furthermore, the ICI therapy has been becoming a main clinical practice for LUAD, alongside surgery, chemotherapy, and targeted therapy. Blockading the immune checkpoints of Programmed Cell Death protein 1 (PD-1) or its ligand PD-L1 is so far the bestdescribed immunotherapy approach and is becoming the routine first-line treatment strategy for NSCLC [8,9]. Although the dramatic therapeutic advantage of ICI treatment was found in both clinical trials and real-world data, a major limitation is that only a subset of patients responds to clinical treatment [10]. Therefore, newly effective determinants to select ESCA patients to receive immunotherapy are urgently necessary.
Age is an important risk factor for most diseases including human tumors. Besides age itself, its relevant molecular traits were also demonstrated to be linked with disease prognosis. Recent studies revealed that particular genes (e.g., APOE and FOXO3), genomic regions (e.g., 5q33.3), and numerous single-nucleotide polymorphisms were associated with longevity [11-15]. It is hard to decompose and explore aging owing to the complex interactions between aging and numerous factors in genome, environment, and age-related diseases [16]. To deeply understand the transcriptome landscape of aging, Peters et al. performed a large-scale transcriptomic exploration and identified aging-relevant genes.
In our work, we collected a total of 351 ESCA samples from 3 independent datasets based on publicly available sources to establish and confirm an aging-relevant risk signature. To explore the possible molecular functions behind the determined risk signature, multi-level immunologic analysis was performed and results revealed the strong capacity of this risk signature for assessing immune microenvironment. Moreover, the identified aging signature could predict ICI therapy response and outcome. Findings derived from this study may provide ideas for survival evaluation and immunogenicity prediction of ESCA patients.
Sample collection and aging-related genes
All eligible ESCA patients with both transcriptomic data and clinical information were acquired from Gene Expression Omnibus (GEO) and the Cancer Genome Atlas (TCGA) projects. Totaling 351 patients were included in this study, they are from TCGA (N=172), GSE53624 (N=119), and GSE53622 (N=60). Among the ESCA cohorts, the TCGA cohort harbored the largest sample size with 172 samples; we thus considered it as the discovery cohort and used it for constructing the risk signature. The detailed cohort features and detection platforms of all ESCA patients were illustrated in Table S1. A total of 348 urothelial cancer patients who received anti-PD-L1 treatment in the IMvigor210 trial were collected to ulteriorly explore the association of the determined aging signature with ICI response and outcome [17]. Totaling 1438 aging-related genes was identified based on a previously published study.
Development of the aging-relevant risk signature
All 1438 aging-relevant genes were performed univariate Cox regression analysis by using transcriptomic profiles of the ESCA discovery dataset to explore the genes with respect to survival status. Then, aging genes associated with survival risk were presented to the Lasso-Cox regression function (achieved by R glmnet package) to identify the genes that contributed most to prognosis [18]. Based on the Lasso regression coefficient results, the optimal gene panel could be determined to establish a risk signature. By integrating particular gene expression values with their corresponding coefficients, risk cores across all ESCA patients could be obtained. The specific calculation formula was: risk score=Coefficient of gene (i) *Expression of gene. ESCA samples were partitioned into low and high-risk subpopulations with median risk score as the cut-off value.
Assessment of tumor infiltrated immunocytes and immune checkpoints
To investigate the different immunocyte infiltration abundance in low and high-risk groups, we calculated the abundance for 28 tumor-infiltrating immunocytes [19]. The 28 immune cells were stratified into three types: anti-tumor, pro-tumor, and intermediate immune cells. The representative genes for each immunocyte type were exhibited (Table S2). CIBERSORT algorithm could evaluate the infiltration levels of 22 immune cell subtypes based on 547 informative genes (termed LM22). In our work, we applied both methods to gain a mutual-confirmation association [20].
A comprehensive integration of immune checkpoint genes was achieved according to a recent immunogenomic research [21]. In the TCGA ESCA cohort, the checkpoint gene of VISTA was not detected due to the distinct sequencing method. We thus calculated the diverse expression of 33 genes in low-versus high-risk groups.
Enrichment of immunogenicity and ICI response indicators
Recent multiple studies have been reported that immune-relevant signatures are associated with immunogenicity and ICI treatment response. Herein, we collected six typical signatures as follows: 1) T cell-inflamed signature; 2) IFNγ signature; 3) Cytolytic activity; 4) immune cell signature; 5) cytokines and chemokines and 6) immune signaling molecules [22-25]. The feature genes applied for assessing enrichment scores of each signature were shown in Table S3.
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) was used to explore signaling pathways enriched in distinct risk subpopulations. The t parameter obtained from the differential analysis achieved by limma package was considered as the input factor for the fgsea method conducted by the R fgsea package [26]. Pathways gleaned from KEGG and HALLMARK databases were used as the comparison signals. Pathway enrichment plot was achieved through the cluster Profiler package [27]. Single sample GSEA (ssGSEA) algorithm embedded in the GSVA package was employed to infer enrichment scores of curated immune signatures and immunocytes for each ESCA patients [28].
Determination of mutational signatures in the genome
We used Signature Analyzer to extract mutational signatures based on somatic mutation data from the TCGA ESCA dataset [29]. Bayesian nonnegative matrix factorization was applied to optimally identify the rank of mutational signatures. Mutation portrait matrix A was divided into two nonnegative matrices W and H (i.e., A ≈ W × H), with W indicating the determined mutational signatures and H reflecting the corresponding mutational activities. The identified mutational signatures were then annotated with the 30 well-curated signatures stored in the COSMIC database based on the cosine similarity [30].
Significantly mutated genes
MutSig CV method was used to determine Significantly Mutated Genes (SMGs) in TCGA ESCA patients [31]. One SMG must meet three criteria: statistically significant, expressed in TCGA ESCA samples and an encyclopedia of cell lines [32]. SMGs mutational patterns were exhibited using the maftools package [33].
Statistical analysis
We employed R software (version 4.1.1) to perform the relevant calculations. This study regarded tumor mutation burden (TMB) as the total nonsynonymous mutation count per megabase. Heatmap illustration of identified aging genes in different risk subgroups was achieved with the pheatmap package. Kaplan-Meier method was used to produce survival plots and the group difference was evaluated by the Log-rank test. Multivariate regression models of the forestmodel package were applied to eliminate the confounding factors and obtain an adjusted association. The correlation of numerical and categorical variables with two risk groups was calculated with Wilcoxon rank-sum test and Fisher exact test, respectively.
Identification of the aging risk signature
Since the ESCA dataset from the TCGA has the largest sample size (N=172) and complete clinical information, we, therefore, considered it as the discovery cohort to construct the aging risk signature. Univariate Cox regression analysis of the collected 1438 aging-relevant genes was performed with transcriptomic data of the TCGA dataset. Results demonstrated that 37 genes were linked with prognosis (all P<0.05; Table S4). We then used the Lasso-Cox regression with 10-fold cross-validation to identify the aging genes that contributed most to the ESCA survival. The Lasso coefficient information between the log (λ) and the gene penal number was shown in Figure 1A. The minimum deviance was observed when the gene number was selected as 22 in Figure 1B. Finally, we chose 22 aging-relevant genes to establish a risk signature for ESCA survival assessment.
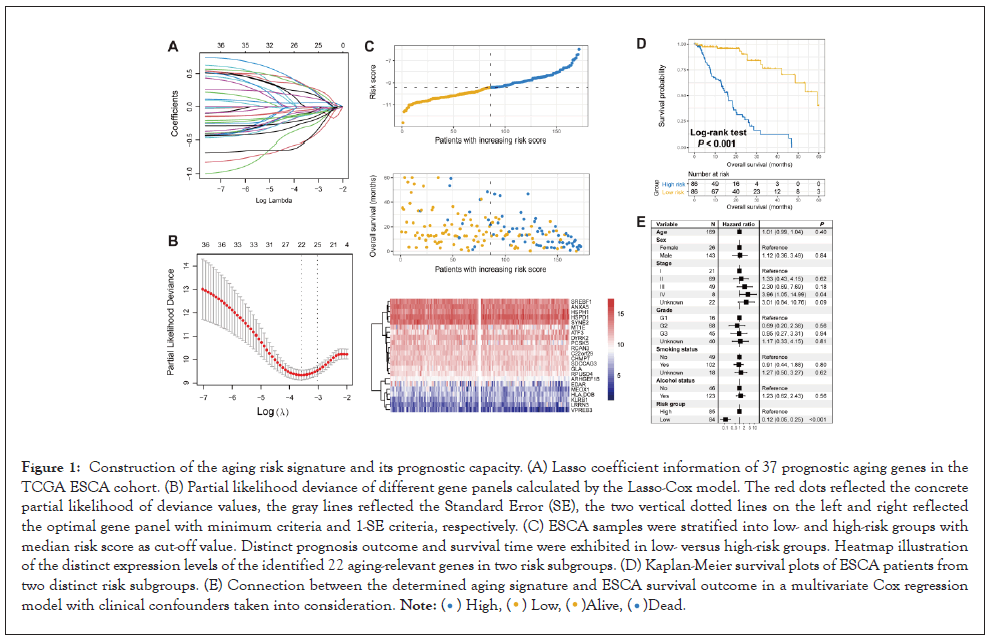
Figure 1: Construction of the aging risk signature and its prognostic capacity. (A) Lasso coefficient information of 37 prognostic aging genes in the TCGA ESCA cohort. (B) Partial likelihood deviance of different gene panels calculated by the Lasso-Cox model. The red dots reflected the concrete partial likelihood of deviance values, the gray lines reflected the Standard Error (SE), the two vertical dotted lines on the left and right reflected the optimal gene panel with minimum criteria and 1-SE criteria, respectively. (C) ESCA samples were stratified into low- and high-risk groups with median risk score as cut-off value. Distinct prognosis outcome and survival time were exhibited in low- versus high-risk groups. Heatmap illustration of the distinct expression levels of the identified 22 aging-relevant genes in two risk subgroups. (D) Kaplan-Meier survival plots of ESCA patients from two distinct risk subgroups. (E) Connection between the determined aging signature and ESCA survival outcome in a multivariate Cox regression model with clinical confounders taken into consideration.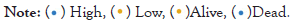 .
.
The determined 22 genes including SDCCAG3, ANXA5, ARHGEF18, VPREB3, MEOX1, RCAN3, EDAR, SYNE2, C22 or f29, PCSK5, CHMP7, RPUSD4, MT1E, HLA-DOB, SREBF1, ATF3, LRRN3, HSPD1, HSPH1, KLRB1, GLA, and DYRK2. Their prognosis contribution values for ESCA patients were exhibited in Table S5. We constructed a risk signature to calculate the risk scores for each ESCA patient in Figure 1C according to the linear combination between the determined 22 gene expression levels and corresponding regression weights. The risk association plot of the calculated risk scores with survival times and statuses was shown in Figure 1C. In addition, distinct expression distribution of the determined 22 genes in low and high-risk groups was also presented with a heat map.
To investigate the survival predictive capacity of the identified risk signature, we stratified ESCA patients from the discovery cohort into low-risk (N=86) and high-risk (N=86) groups. We found that patients of low-risk group had a significantly better overall survival than patients of high-risk group (Log-rank test P<0.001; Figure 1D). This link was still significant after controlling for age, sex, stage, grade, smoking status, and alcohol status in a multivariate Cox regression model (HR: 0.1295% CI: 0.05-0.25, P<0.001; Figure 1E) (Figures 1A-1E).
Corroboration of the aging risk signature
To validate the prognosis predictive roles of the constructed risk signature, we used the Disease-Specific Survival (DSS) and Progression-Free Survival (PFS) information from the TCGA ESCA cohort. Besides, two additional ESCA cohorts derived from the GEO were also used for validation. We observed that ESCA patients with a low-risk score exhibited both improved DSS and PSS as compared with high-risk patients in the TCGA cohort (Logrank test both P<0.001; Figures 2A and 2C).
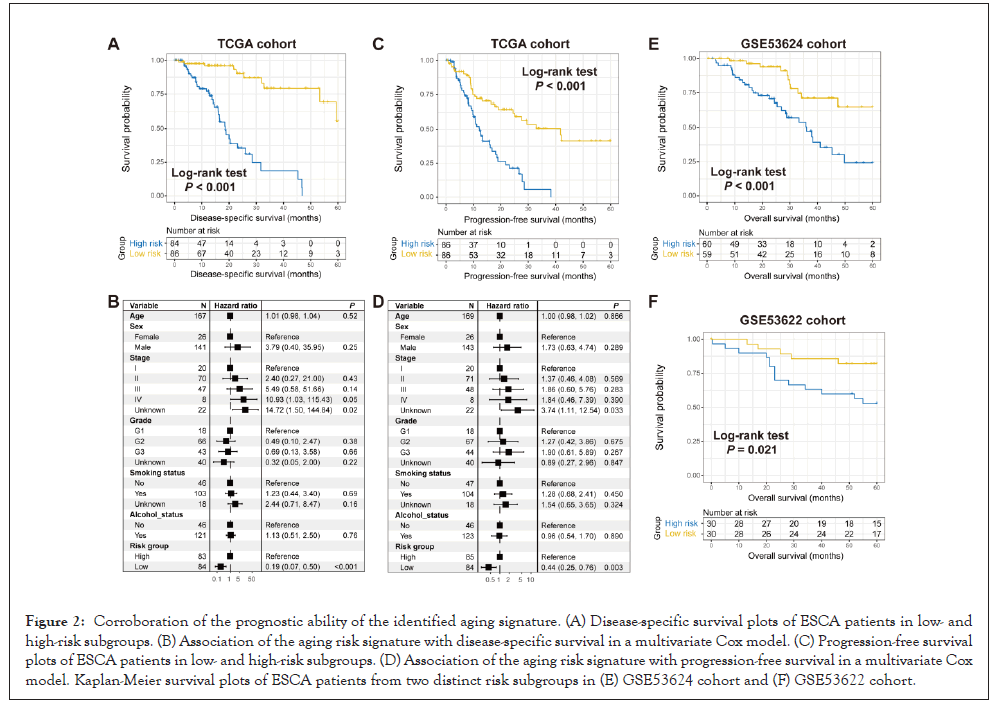
Figure 2: Corroboration of the prognostic ability of the identified aging signature. (A) Disease-specific survival plots of ESCA patients in low- and high-risk subgroups. (B) Association of the aging risk signature with disease-specific survival in a multivariate Cox model. (C) Progression-free survival plots of ESCA patients in low- and high-risk subgroups. (D) Association of the aging risk signature with progression-free survival in a multivariate Cox model. Kaplan-Meier survival plots of ESCA patients from two distinct risk subgroups in (E) GSE53624 cohort and (F) GSE53622 cohort.
The associations remained still significant when controlling for clinical confounding factors (multivariate Cox regression P<0.001 and P=0.003, respectively; Figures 1B and 1D). In two GEO cohorts of GSE53624 and GSE53622, the significantly prolonged overall survival outcomes were also observed in patients with low-risk scores in Log-rank test P<0.001 and P=0.021, respectively; Figures 2E and 2F (Figures 2A-2F).
The identified risk signature associated with preferable immunocyte infiltration and immune activity
Previous two studies have been demonstrated that aging and its relevant genomic traits were involved in the immune microenvironment and immunity [34,35]. Hence, we speculated that the determined aging signature might regulate the immune cell infiltration and biological circuits associated with the immune activity. A heatmap according to ssGSEA algorithm was achieved to show the different infiltrated levels of 28 immunocyte types in lowversus high-risk groups in the TCGA ESCA cohort in Figure 3A. Results revealed that increased infiltration of anti-tumor immune cell types, such as central memory CD4+ and CD8+ T cells, and effector memory CD8+ T cells were observed in ESCA patients of low-risk group (all P<0.05). Moreover, the decreased infiltration of pro-tumor immunocytes, like neutrophils and regulatory T cells were noticed in the low-risk group (both P<0.05). In addition, activated B cells and gamma delta T cells, which belong to the intermediate immune cell type, were also markedly enriched in patients with low-risk scores (both P<0.05). We also performed immunocyte infiltration analysis via the CIBERSORT method in Figure S1, and consistently, more infiltration of immune response cells (Ex: CD8 T cells, activated natural killer cells, and M1 macrophages) and less infiltration of immune suppressive cells (Ex: regulatory T cells and M2 macrophages) were observed in low-risk ESCA patients.
Figure 3: Connection of the aging signature with immune infiltration and immunogenicity. (A) Infiltration levels of different immunocytes in low- and high-risk ESCA groups. Immunocyte highlighted with red suggested its infiltration was markedly enhanced in the low-risk group, while the blue suggested the infiltration was markedly decreased in the low-risk group. GSEA pathway analysis of low-risk patients with comparison circuits from (B) KEGG and (C) HALLMARK databases, respectively. Pathways marked with green were immune response-relevant pathways. Different enrichment of (D) T cell-inflamed signature, (E) IFNγ signature, and (F) cytolytic activity signature in low- versus high-risk ESCA patients. *P<0.05, **P<0.01. Note: Risk group: 
 .
.
GSEA analysis against KEGG and HALLMARK databases was conducted to investigate the specific biological pathways involved in the aging signature. We found that immune response-relevant circuits in the KEGG database (Ex: T cell receptor signaling pathway and chemokine signaling pathway) and HALLMARK database (Ex: Inflammatory response, interferon-gamma response, and IL2-STAT5 signaling) were markedly enriched in the ESCA low-risk subgroup in Figures 3B and 3C.
ESCA patients of low-risk group had a significantly enhanced enrichment of T cell-inflamed signature, IFNγ signature, and cytolytic activity than those patients of high-risk group (Wilcoxon test all P<0.05; Figures 3D-3F). Besides, the elevated enrichment of immune cell signature, immune signaling molecules, and cytokines/chemokines signature was also observed in low-risk patients (Wilcoxon test all P<0.05; Figure S2) (Figures 3A-3F).
The different expression levels of complete immune checkpoints between low and high-risk subpopulations were calculated. Results showed that the common immune checkpoints (e.g., CD274, CD276, IDO1, LAG3, and PDCD1) were highly expressed in patients with a low-risk score (all P<0.05; Figure S3).
TMB and SMGs linked with the determined aging signature
Tumor mutation burden (TMB) was revealed to be associated with tumor prognosis and immunotherapy response [36,37]. We thus explored the connection between the aging signature and TMB. By using the somatic mutational profile of TCGA ESCA dataset, we calculated the TMB for each ESCA patient and noticed that patients with a low-risk score harbored a markedly enhanced TMB than patients with a high-risk score in Wilcoxon test, P<0.001; (Figure 4A).
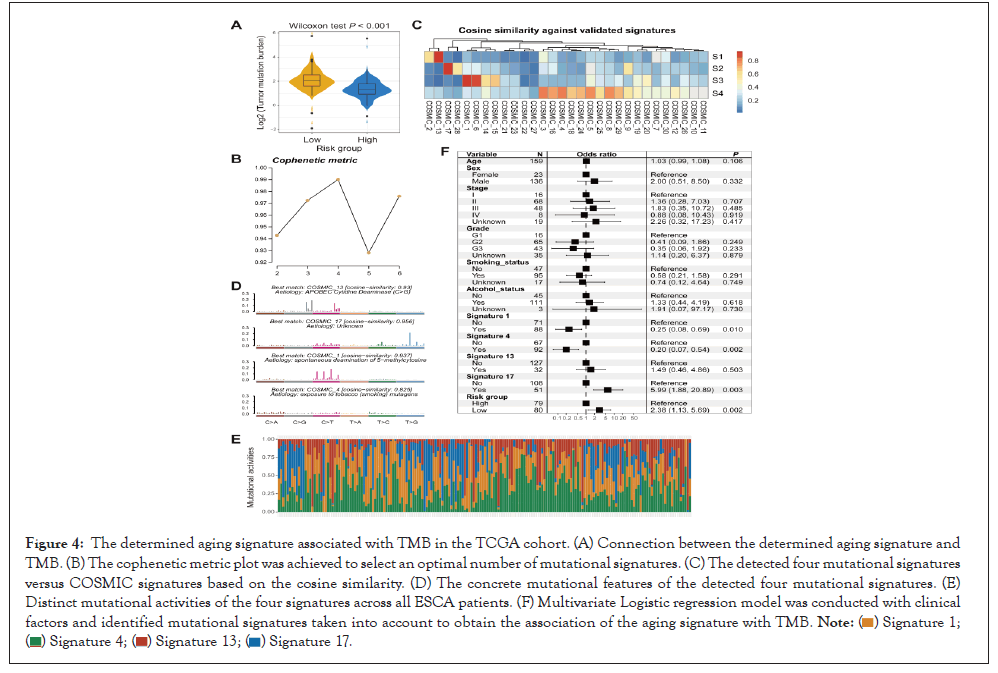
Figure 4: The determined aging signature associated with TMB in the TCGA cohort. (A) Connection between the determined aging signature and TMB. (B) The cophenetic metric plot was achieved to select an optimal number of mutational signatures. (C) The detected four mutational signatures versus COSMIC signatures based on the cosine similarity. (D) The concrete mutational features of the detected four mutational signatures. (E) Distinct mutational activities of the four signatures across all ESCA patients. (F) Multivariate Logistic regression model was conducted with clinical factors and identified mutational signatures taken into account to obtain the association of the aging signature with TMB.
 .
.
Mutational signatures are characterized by specific combination patterns of nucleotide substitutions and have been demonstrated to associate with TMB. Hence, we detected possible mutational signatures of ESCA samples by dividing the nucleotide substitution matrix under the NMF approach. Based on the cophenetic metric plot, four mutational signatures were determined in Figure 4B.
After annotating them with signatures from the COSMIC database in Figure 4C, we identified these four mutational signatures as signature 1 (associated with age of cancer diagnosis), signature 4 (associated with smoking), signature 13 (associated with APOBEC enzyme activity), and signature 17 (the etiology of this signature remains unknown). Distinct mutational features of the above four signatures were exhibited in Figure 4D. Mutational signature activity of all ESCA samples was calculated and illustrated in Figure 4E and Table S6. Subsequent investigation indicated that low-risk scores were significantly associated with the elevated mutational activity of signature 1 (P=0.036; Figure S4A) and decreased activity of signature 17 (P=0.049; Figure S4B). No significant differences were found between two risk groups concerning signatures 4 and 13 (P=0.829 and 0.779, respectively).
To elucidate whether the enhanced TMB of low-risk group was affected by other clinical confounders, we added clinical features (i.e., age, sex, stage, smoking status, and alcohol status) and determined mutational signatures (i.e., signatures 1, 4, 13, and 17) into a multivariate logistic regression model. The connection between the low-risk scores and the elevated TMB was still remained (OR: 2.38, 95% CI: 1.13-5.69, P=0.002; Figure 4F) (Figures 4A-4F).
Totaling 16 significantly mutated genes (SMGs) were determined by detecting the somatic mutation data of the TCGA cohort. Waterfall plot between low and high-risk groups demonstrated a markedly different mutation frequency in TP53 [66 of 80 (82.5%) vs. 71 of 79 (89.9%); P=0.038], NAV3 [3 of 80 (3.8%) vs. 11 of 79 (13.9%); P=0.025], and FAT1 [2 of 80 (2.5%) vs. 6 of 79 (7.6%); P=0.045] (Figure 5).
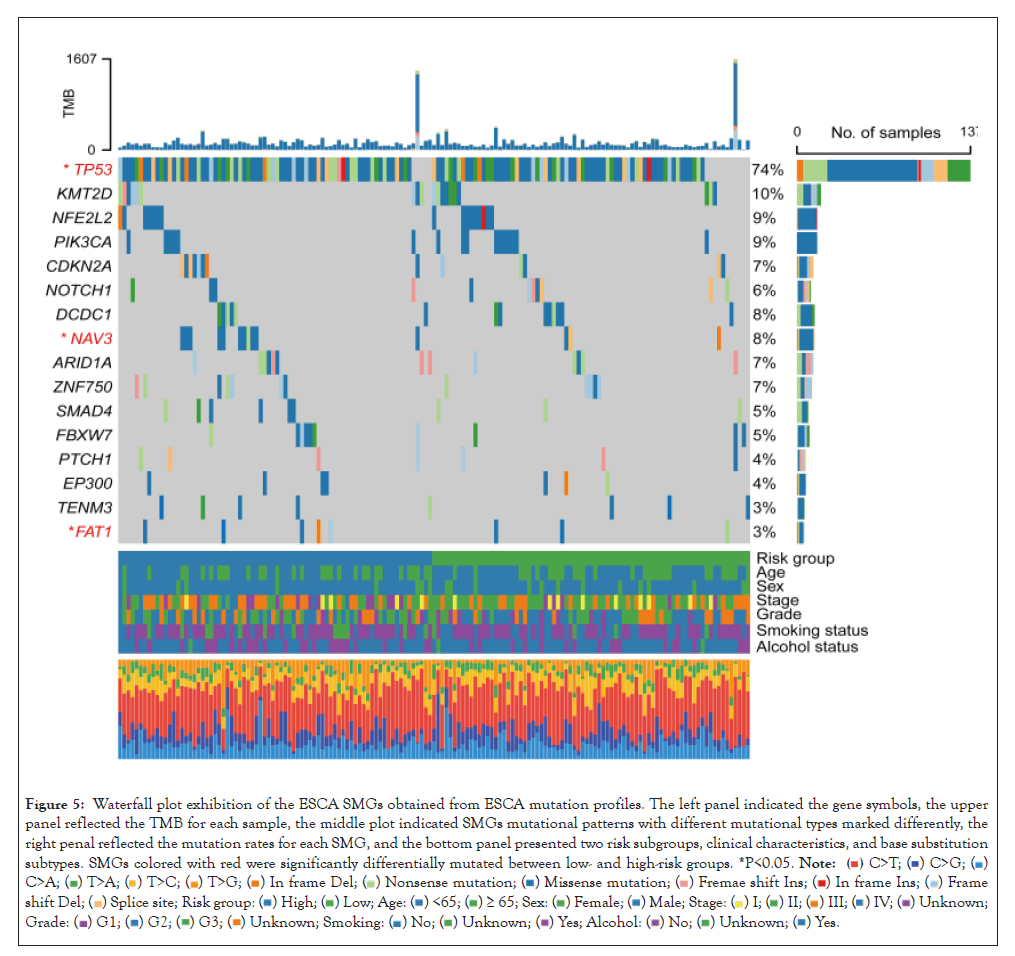
Figure 5: Waterfall plot exhibition of the ESCA SMGs obtained from ESCA mutation profiles. The left panel indicated the gene symbols, the upper panel reflected the TMB for each sample, the middle plot indicated SMGs mutational patterns with different mutational types marked differently, the right penal reflected the mutation rates for each SMG, and the bottom panel presented two risk subgroups, clinical characteristics, and base substitution subtypes. SMGs colored with red were significantly differentially mutated between low- and high-risk groups. *P<0.05.

Roles of the determined aging signature in assessing ICI treatment efficacy
The aforementioned results demonstrated that the established aging risk signature was connected with immune microenvironment and immunogenicity; we hypothesized that the aging signature may play vital roles in immune checkpoint inhibitor (ICI) treatment. Hence, an urothelial cancer anti-PD-L1 dataset (Imvigor210) with both gene expression profiles and immunotherapeutic information was used to investigate the connection between determined aging signature and ICI efficacy. Results revealed that patients of low-risk subgroup had a markedly better ICI prognosis as compared with patients of high-risk subgroup (Log-rank test P=0.002; Figure 6A). This association was remained even controlling for sex, baseline ECOG score, smoking status, immune phenotype, and platinum treatment in a multivariate Cox regression model (HR: 0.69, 95% CI: 0.53-0.91, P=0.007; Figure 6B). Subsequent analysis showed that a higher proportion of better ICI response status (i.e., CR and PR) was observed in the low-risk group (28.7% vs. 16.9%, P=0.018; Figure 6C). Besides, in this ICI-treated cohort, we also observed patients with low-risk scores exhibited an elevated TMB (Wilcoxon test P=0.004; Figure 6D). A trend of higher neoantigen burden was also noticed in the low-risk subgroup, although this association did not reach statistical significance (Wilcoxon test P=0.083; Figure 6E) (Figures 6A-6E).
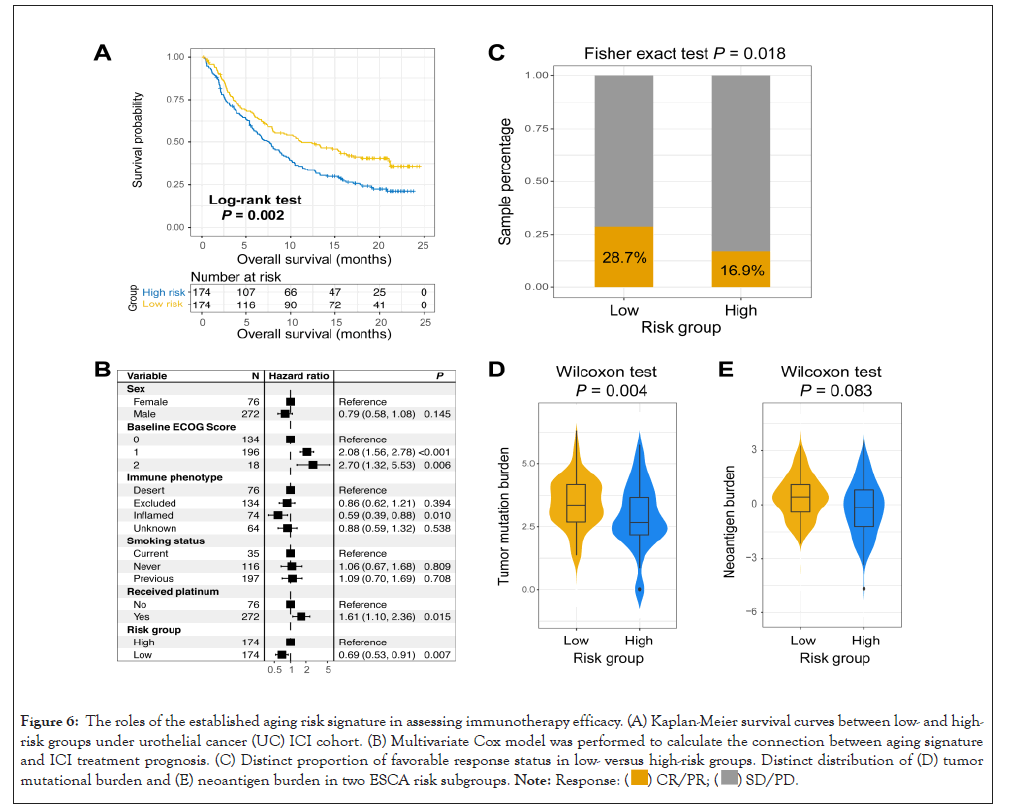
Figure 6: The roles of the established aging risk signature in assessing immunotherapy efficacy. (A) Kaplan-Meier survival curves between low- and high-risk groups under urothelial cancer (UC) ICI cohort. (B) Multivariate Cox model was performed to calculate the connection between aging signature and ICI treatment prognosis. (C) Distinct proportion of favorable response status in low- versus high-risk groups. Distinct distribution of (D) tumor mutational burden and (E) neoantigen burden in two ESCA risk subgroups. .
.
Finally, we conducted immune infiltration analysis by using transcriptomic data of this immunotherapeutic cohort to compare the distinct infiltration levels between two risk subgroups. Consistently, the enhanced infiltration of immune response lymphocytes represented by CD8 T cells was significantly enriched in patients with low-risk signature scores in Figure S5.
In this work, by using gene expression profiles and clinical data from several ESCA datasets, we constructed and validated an agingrelevant risk signature for prognosis evaluation, immunogenicity, and ICI response prediction. Future prospective studies are necessary, but findings obtained from our research suggest the potential roles of the aging risk signature in ESCA clinical prognosis and immunotherapeutic efficacy surveillance.
Mutational signatures are the manifestations of endogenous and exogenous factors that indicated specific mutational patterns [38]. Amongst, the aging-relevant mutational signature 1 was revealed to be linked with the poor tumor microenvironment and prognosis in triple-negative breast cancer, indicating its possible roles for immune treatment efficacy [39]. Subsequently, a recent study integrated somatic mutational profiles of melanoma and NSCLC samples that received ICI therapy and noticed that both tumors with aging signature showed a worse survival outcome [40]. In our study, different from mutation-level biomarkers, an aging-relevant risk signature was constructed at the transcriptomic level, and we observed this signature harbors the ability to predict prognosis outcome and immunogenicity in ESCA.
Of the identified 22 aging genes, six (i.e., ANXA5, MEOX1, PCSK5, HLA-DOB, SREBF1, and KLRB1) were also demonstrated to be associated with tumor immunity. ANXA5 was associated with infiltration of antigen-presenting cells in glioma and was recently identified as a novel immune checkpoint inhibitor [41,42]. A molecular signature contained MEOX1 was constructed to predict survival and immune immunologic status in breast cancer [43]. PCSK5 was linked with cancer alternative splicing events, which contribute to carcinogenesis and immune microenvironment in head and neck squamous cell carcinoma [44]. In ovarian cancer, an immune-related prognostic model including HLA-DOB was determined to be connected with prognosis and immunity [45]. Enhanced transcription of SREBF1 promoted invariant natural killer T cell activity, thus increasing lipid biosynthesis and inhibiting anti-tumor effect [46]. A recent study reported that loss of function of KLRB1 enhanced T cell-mediated toxicity and anti-tumor function in glioma [47]. The above evidence further confirms the potential implications of established risk signature in immune infiltration and immunotherapy efficacy.
TP53, NAV3, and FAT1 are frequently mutated in ESCA and their mutations are associated with an inferior survival outcome in ESCA and other several cancers [48-51]. In this study, the relatively lower mutation frequency of the above SMGs was enriched in the ESCA low-risk subgroup, which is consistent with the finding that patients of low-risk group exhibited a favorable prognosis. However, recent studies have also reported that TP53 mutations could predict a better efficacy in cancer immunotherapy; this suggests that lesser TP53 mutations in the low-risk ESCA patients contribute to the survival benefits rather than immunotherapy efficacy. Future indepth studies are warranted [52].
Given the lack of ESCA datasets with both gene expression profiles and ICI treatment data, we thus used an urothelial cancer immunogenomic dataset [17], which is so far the largest immunotherapy dataset to explore the connection of the aging risk signature with ICI treatment efficacy. Results demonstrated the preferable ICI prognosis and response status (i.e.CR and PR) were presented in patients of low-risk group. Considering the cancer homogeneity in particular situations (e.g., therapy outcome assessment), we conclude that the aging-relevant signature is predictive of ICI efficacy not only in urothelial cancer but also ESCA and other tumor subtypes.
Limitations exist in our study. First, the gene expression profiles of ESCA samples were acquired from publicly available datasets, which might produce deviation in the analysis procedure of different cohorts. Second, relevant results from the genomic mutational profile were calculated only based on the TCGA ESCA dataset, no additional mutational data was used for validation. Finally, experimental verification used for multiple associations is lacking.
In summary, by employing the aging gene expression data of ESCA samples, we constructed a risk prediction model to evaluate prognosis, immunogenicity, and immunotherapy response. The novel determined signature might be a possible indicator for ESCA clinical monitoring and treatment.
None
Due to the nature of this study, no ethical approval was required.
The authors certify that there are no conflicts of interest regarding this manuscript.
None
JY and JZ conceived this study; WD, WF, JQ, and HC collected the related data and performed main data analysis; WD, WF, JQ, and GJ conducted data analysis and interpretation; WD, WF, and JQ drafted and corrected the paper; JY and JZ supervised this study.
All genomic data and clinical information employed in this work are obtained from publicly accessible dataset.
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
Citation: Cheng H, Huang C, Qiu H, Zhang W, Chen L, Song X, et al. (2022) Characterization of Aging-Related Profiles in Esophageal Cancer Reveals a Signature Predicting Favorable Outcome and Immunogenicity. Chemo Open Access 10:158.
Received: 30-Mar-2022, Manuscript No. CMT-22-17003; Editor assigned: 01-Apr-2022, Pre QC No. CMT-22-17003 (PQ); Reviewed: 13-Apr-2022, QC No. CMT-22-17003; Revised: 21-Apr-2022, Manuscript No. CMT-22-17003 (R); Published: 02-May-2022 , DOI: 10.4172/2167-7700.22.10.158
Copyright: © 2022 Cheng H et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.