Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Review Article - (2023)Volume 16, Issue 3
Background: New molecular docking software is a technical approach that is mostly employed in drug discovery and design, as well as for numerous analysis processes. Computerized platforms are used in the process of molecular docking. It is a molecular combination in which one molecule binds to other molecules to form a bonded complex with all-around lowest energy requirements.
Cannabidiol (CBD) has blowup in fame around the world in just a few years. Cannabidol is currently vended and used to give a wide range of therapeutic illnesses and lifestyle diseases after being revealed as an effective selfmedication for Dravet syndrome in progenies. Cannabidol, a non-psychoactive isomer of the more well-known Tetrahydrocannabinol (THC), is becoming increasingly widely accessible in a variability of forms.
Result: Using several molecular docking techniques, the current study seeks to investigate the binding interactions of delta-9 THC with cytochrome p450. CYPs are a family of mono oxygenases that contain heme as a cofactor. The findings of docking tests revealed that these compounds have high binding affinities. The mechanism and interactions are the focus of this research.
Conclusion: It aids computer-based concepts, which provide new ideas to staff in investigating and bringing theories together experimentally, as well as bringing answers to all difficulties, including cost, that arise.
Cytochrome p450; Delta-9 THC; Molecular docking; Binding interactions; Narcotics
THC: Delta-9 Tetrahydrocannabinol; MD: Molecular Docking; CYPs: Cytochrome P450
A flowering plant genus belonging to the Cannabaceae family is Cannabis. It is unknown how many species there are in the genus [1]. The three types that can be distinguished are Cannabis sativa, Cannabis indica, and Cannabis ruderalis. Cannabis ruderalis could be considered a subspecies of Cannabis sativa, and all three could be considered one species. The genus is commonly acknowledged as being native to and initiating in Central Asia, with other studies also claiming that it originated in upper South Asia [2].
Plants have been used as medicine for over 8000 years, according to a large body of literature. The first documented text on the medicinal use of Cannabis was discovered in China. Between 1200 B.C. and 800 B.C., the Atharvaveda, Hinduisms sacred literature, cites the psychoactive qualities of Cannabis and its use for therapeutic purposes in India [3].
Delta-9 Tetrahydrocannabinol (THC), a cannabinoid found in both male and female plants, is the major active principle. THC is found in the highest quantity in the bracts, flowers, and leaves, whereas it is almost non-existent in the stem, root, and seeds (Figure 1).
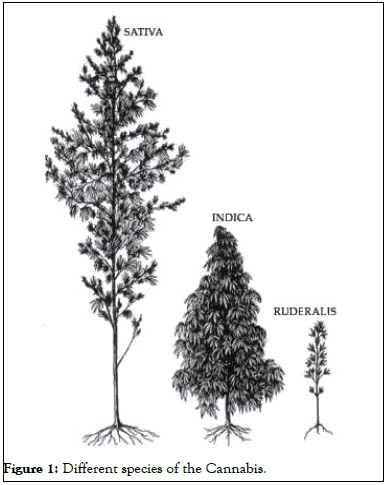
Figure 1: Different species of the Cannabis.
The amount of THC in a plant varies a lot, and it is probably influenced more by the seed type than by soil or climatic conditions. cannabidiol, cannabinol, cannabidolic acid, cannabicyclol, and cannabigerol are among the cannabinoids found in Cannabis sativa, in addition to THC. More than 60 cannabinoids have been identified thus far (Figure 2).
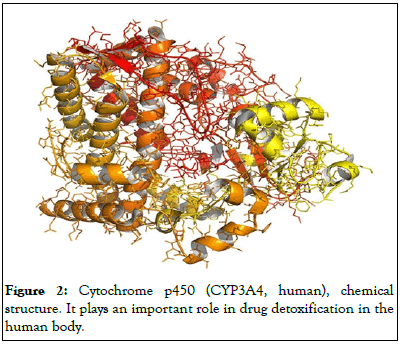
Figure 2: Cytochrome p450 (CYP3A4, human), chemical structure. It plays an important role in drug detoxification in the human body.
Cannabis is an antidepressant. Antidepressant medicines do not always cause depression. They slow down the central nervous system, movement, and the transmission of signals from the brain to the body relative to other factors. When substantial quantities of cannabis are taken by any individual, it can cause hallucinatory effects (Figure 3).
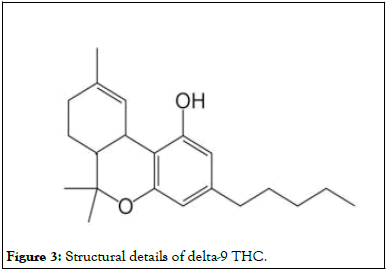
Figure 3: Structural details of delta-9 THC.
Cannabis, commonly referred to as marijuana, is a psychoactive material derivative from the cannabis plant that is used for therapeutic purposes. Cannabis can be used in a various activities, involving smoking, vaporizing, ingesting it in food, and extracting it. Cannabis is primarily used for recreational or healing purposes, while it can also be utilized for spiritual motives (Figure 4).

Figure 4: It is the ribbon like representation of the cytochrome p450 in the discovery studio visualizer.
THC is absorbed into the bloodstream through the lungs or stomach and intestines walls. THC travels through the bloodstream to the brain, where it produces high effects. Inhaled drugs reach the bloodstream faster than those consumed. This means that when Cannabis is smoked, the effects are felt faster than when it is ingested (Figure 5).
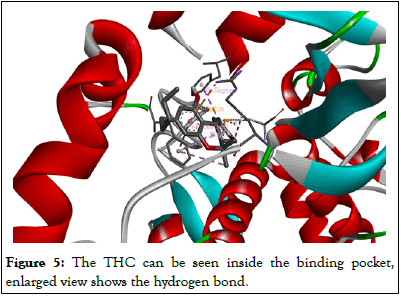
Figure 5: The THC can be seen inside the binding pocket, enlarged view shows the hydrogen bond.
Cannabis has both psychological and physical impacts. THC and its primary metabolite, THC-COOH, can be identified using chromatographic techniques in blood, urine, hair, oral fluid, or sweat as quantity of a drug-testing package or a forensic inquiry into traffic or another illegal crime. The concentrations acquired from such examines can frequently be useful in determining active versus passive revelation, as well as the amount of time since usage and the intensity or length of use.
Receptor
Receptors are proteins that recognize a molecule or a group of chemicals (ligands) with enough specificity to trigger a signaling cascade that connects to secondary messenger systems (Figure 6).
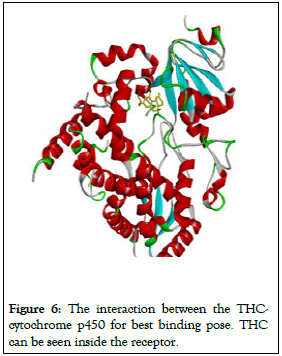
Figure 6: The interaction between the THC-cytochrome p450 for best binding pose. THC can be seen inside the receptor.
There are many different types of receptors, they are split in two different groups: Intracellular receptors present inside the cell and cell surface receptors present on the plasma membrane.
Receptors are proteins that play an important role in the body’s communication system. The vast majority of these are found within the cell membrane [4].
Receptor proteins are implanted in the cell membrane in such a way that their active site-containing tiny component protrudes from the membranes surface and opens onto the cell membranes the outside region (Table 1).
| Mode | Affinity (kcal/mol) | Dist. from rmsd 1.b. | Best mode.rmsd u.b. |
|---|---|---|---|
| 1 | -9.2 | 0 | 0 |
| 2 | -8.9 | 2.212 | 6.18 |
| 3 | -8 | 2.796 | 6.67 |
| 4 | -8 | 3.128 | 5.747 |
| 5 | -7.5 | 5.387 | 8.373 |
| 6 | -7.1 | 8.639 | 11.879 |
| 7 | -7 | 20.931 | 24.045 |
| 8 | -6.9 | 10.29 | 11.049 |
| 9 | -6.9 | 8.827 | 10.577 |
Table 1: Affinity of receptors.
The body contains a huge number of distinct receptors that interact with various chemical messengers. Because their binding sites are different in shape, structure, and amino acid makeup, these receptors demonstrate selectivity for one chemical messenger over the other. Antagonists are drugs that attach to the receptor site and prevent it from performing its normal action [5]. When message blocking is required, these are useful. Other medications, known as agonists, replicate the natural messenger by turning on the receptor. They are useful when a natural chemical messenger is not available [6].
When any ligand attaches to the appropriate location on the receptor, it generally operates via amplification, integration, or relaying of signals [7]. Although a cell contains hundreds of identified receptors, they tend to identify and bind to only one type of ligand, resulting in activation or inhibitory processes.
The cytochrome p450 receptor is the subject of this research.
Cytochrome p450
Cytochrome p450 is a monooxygenase enzyme family that contains heme as a cofactor [8]. Steroids, fatty acids, and xenobiotic are oxidized by these proteins in mammals, and they are vital for the clearance of different substances, as well as hormone creation and cessation [9]. These proteins are required in the manufacture of defense chemicals, fatty acids, and hormones in plants. A heme-iron center is found in the active site of cytochrome p450.
The name cytochrome p450 enzymes come from the fact that they are attached to cell membranes and contain heme pigment, among other things [10].
These proteins emit a spectrum with a wavelength of around 450 nm when attached to carbon monoxide.
Human cytochromes are mainly membrane-associated proteins found in mitochondrial inner membranes of the endoplasmic reticulum of cells. Many medicines can either raise or reduce the activity of CYP isozymes by affecting their production or directly blocking their function [11].
Ligand
An ion or small molecule known as a ligand attaches to a metal atom in the center to form a complex. Ligands can take many various forms and interact with receptors on or in target cells, are created by signaling cells. When ligand binds to the receptor, it can trigger certain signals, which helps to initiate or inhibit the biological processes [12]. It is observe that the binding of the ligand and receptors is generally due to the weak forces such as hydrogen bonds, van der waal forces and ionic bonds [13]. Ligand can be classified into the different types according to the biological processes they regulate such as substrate, inhibitors, activators and neurotransmitters [14].
Present study deals with the cannabinoids as ligand, which includes delta-9 THC. Cannabinoid are a huge group of active compounds found in plant Cannabis.
Δ-9-THC
THC, also known as Dronabinol is the key component responsible for the pharmacological and psychotropic effect caused by Cannabis species. It is one of at least 113 cannabinoids recognized in Cannabis. THC is the psychoactive constituent of Cannabis. THC shows its rapid absorption from lungs to blood, when smoked. Being lipophilic in nature, it is distributed rapidly throughout the tissues and later to fatty layers [15]. THC interacts with various neuromodulators and neurotransmitters. It may sometimes induce or suppress pain, convulsions or tremors depending upon the condition [16,17].
Molecular docking Docking is a structure-based technique for determining the optimum fit between two molecules [18]. Docking is a system in molecular modeling that calculates the preferred orientation of one molecule to another when they are linked together to make a stable compound. The strength of binding affinity between two molecules can be foretold using information about the preferred orientation [19]. The primary situation for performing a docking screen is the structure of the protein. The structure is usually resolute using a biophysical technique like x-ray crystallography or nuclear magnetic resonance spectroscopy, although it can also come through homology modeling [20]. A docking device uses this protein structure and a databank of possible ligands as inputs. A docking programs success is determined by two factors: The search algorithm and the scoring mechanism. The goal of molecular docking is to effectively forecast the monovalent interaction of macromolecules and a small molecule (ligand) starting from their unbound structures, structures obtained from MD simulations or homology modeling, etc. Predicting bound conformations and binding affinity is the goal. The operation of these tools is divided into four stages. The first involves determining the structure using various techniques such as NMR22. The production of the receptor and ligand files is the second phase, and the computational procedure employing various algorithms is the third. The analysis of the results obtained by docking is the final phase.
Molecular docking is a technique for analyzing the three-dimensional information of structures (receptors and ligands) and predicting various binding modes and places. Structural molecular biology and computer-assisted drug design, it is a crucial tools. The goal of docking is to anticipate the most typical ways that a ligand will attach to a protein with a known three-dimensional structural structure. Docking can be used in lead optimization to do virtual screening on huge libraries of complexes, analyses the results, and offer structural suggestions for how the ligands inhibit the target.
It can be challenging to sometimes take the outcomes of stochastic search processes into account because the input configurations for docking are just as important as the docking itself. Competent high-dimensional space exploration and an exact docking candidate rating system are used by effective docking algorithms.
Kolb et al., employed used molecular docking to screen a library of about a million readily available lead-like structures for commercial use. Computer-Aided Drug Design and Discovery (CADD) has grown in popularity during the past few years. CADD is used by many large pharmaceutical companies for lead discovery. The process is challenging, and it is challenging to manage the interactions that take place, hence enhancing molecular docking modelling has been important in many situations for several operations.
Damm-Ganamet et al., published a structured activity resource document for the community in the year 2013. Even though many docking programs have been organized over the last two decades, it is still challenging to grasp docking programs for specific targets.
Emil Fischer had a good year. The well-known "lock and key system," which serves as a model for explaining, was the first discovery of explanation. Crosland introduced the "induce fit" technique in 1958. The enzyme-substrate interaction, according to this idea, causes conformational alterations that optimize ligand-target interactions. The landscape for energy, which provides the work for the protein confirmations that a ligand attaches to them preferentially, was described in the year 1965, and one evaluation was established in the year 2011.
Aim and objective
The basic goal of docking is to determine the ideal manner for a ligand to connect to an active site to obtain optimal conformational for both the receptor and the ligand.
• To accomplish protein-ligand orientation with the least
amount of energy expended.
• Its primary function is to identify the correct binding site for
the protein.
Computational tools used for the study
Autodock vina (VERSION 1.5.6): Autodock vina was developed at molecular graphics lab (The Scripps research institute) by Dr. Oleg Trott. This software is an open source and is extensively used for molecular docking. Autodock vina is known for reduced running time due to use of multiple CPU of the system and is easier for user to operate. This software uses scoring functions which help in predicting the non-covalent interactions and binding affinities between the receptor and the ligand. It produces all the possible poses a molecule attains during the docking procedures.
Discovery Studio Visualizer (DSV): BIOVIA DS visualizer is a suit of freeware developed by Accelrys. It is used for simulation and molecular modeling of various bio molecules. This program has wide range of application in life sciences, which allows user to analyze and visualize the bio chemical data. It allow user to view three dimensional molecules in different modes. Discovery studio is also used to view results obtained by various docking tools such as autodock vina.
Pymol: Warren Lyford Delano designed the computer software, which is a molecular visualization system. It is open source software that been funded by users and released under the Python license. The first to commercialize it was Delano scientific LLC, a private software company devoted to offering precious apparatus that are typically accessible to scientific and educational groups. Schrodinger, Inc. is now marketing it. PyMol can generate high-resolution three-dimensional images of tiny compounds as well as biological macromolecules like proteins. PyMol was used to produce around 25% of all published images of three-dimensional protein structures in the scientific literature by 2009. It is one of the few open-source model visualization apparatuses for structural biology that are easily accessible.
Structural investigations: Present study was carried out using the different computational tools. For this study data of the receptor and ligand is made available on Protein Data Bank (PDB). Most of the tools used for read and recognize the files in PDB or PDBQT format. In case of unavailability of PDB files, programs like open babel can be used to convert any file into the PDB format.
Collection of data bank: The necessary PDB files for carrying out computational studies can be made available at RCSB protein data bank, a worldwide database for the structural data of bio molecules. This is an online database and allows users to download the files in PDB format.
Docking process steps: The vital phases which are used in a docking procedure are as follows:
A directory has to be created for saving all the files and initially contains the receptor file, ligand file and configuration file in PDBQT format. Once the run is complete the log file and output files are also saved in the same.
Creation of target (protein)
• Open auto dock tools.
• File, read molecule, open.
• Edit, hydrogen, add, polar only, remove water molecules
(if required).
• Grid, macromolecule, choose, select molecule (save in
the same folder in PDBQT format).
• Setting the grid box size-go to tools again, grid, grid box,
adjust spacing if required, center grid box (by changing
x, y, z, center), reduce or increase the box size in x, y, z
directions around the active site, note down all box
parameters, close grid dialog box.
Formation of ligand file
• Ligand, input, open.
• Display, show/hide molecule, receptor; on/off.
• Ligand, torsion free, choose torsion, done.
• Ligand, output, save as pdbqt.
Making of instruction file
Generate a file named conf.txt for autodock vina in notepad.
Docking calculations
The receptor and the ligand files are saves in the PDBQT format along with the conf.txt file in the same working directory with the use of command prompt and then the docking calculations are performed.
Open the command prompt and go to the working directory by typing the name of the directory and then start the docking calculations by typing the commands such as;
Results are obtained as log.txt file and the changes in the structural configuration of the ligand are obtained as out.pdbqt. Output files basically show the different poses and also the affinity values.
Autodock vina provides the binding affinities of different poses arranged in the descending order as log.txt file. The changes in the output file of ligand after docking or we can say that the results can be analyzed in the PyMol. It allows to observe the three dimensional molecules in the different modes and save the viewing results according the type of file required.
Molecular docking was carried out between the one receptor and one ligand. The value of exhaustiveness for the calculation was 8 and the grid parameters are the following:
• Center x-1.00, center y-1.00, center z-1.00
• Size x- 98, size y-120, size z-104
The interaction of ligands with the receptor is influenced by the structural modifications in the ligand and the binding sites of the receptor. Ligands having the certain areas in their structure through which they can interact with the receptor by forming the hydrogen bonds or van der waal interactions.
The cytochrome p450 interactions
The results obtained as log.txt file and output file shows the nine binding poses for THC-Cytochrome p450 interactions, with best pose having the affinity of -9.2 kcal/mol. lower the binding energy more stable is the complex.
QM-MM approach
Molecular mechanics is a computational technique that uses potential functions derived from classical physics to compute the potential energy surface for a specific arrangement of atoms. The force field potential of Molecular Mechanics (MM) is used to characterize the ligand and receptor interactions in this method.
An inter-atomic potential fundamental functional form incorporates both bonded and non-bonded terms referring to atoms linked by covalent bonds. Although the specific decomposition of the terms varies on the force field, the total energy can be expressed as;
Etotal=Ebonded+Enon-bonded
Where the constituents of the covalent and non-covalent contributions are given by the following summations;
Ebonded=Ebond+Eangle+Edihedral
Enon-bonded=Eelectrostatic+Evan der waals
The molecular mechanics energy equation, which is a sum of elements, is used to determine the energy due to bond stretching, angle bending, torsional angles, hydrogen bonds, Van Der Waals forces, and columbic attraction and repulsion.
Vtotal=Σangles Vbond+Σangles Vangle+Σdihed Vdihed+Σimpr Vimpr+Σ(i≠J) (VvdW+Velec)
Quantum mechanics computations, on the other hand, are time expensive and are commonly utilized to research chemical reaction mechanisms. It is an extremely precise tool for analyzing transitional stages. It is quite expensive, and it is only good for very small systems with high accuracy, like DFT. It can be utilized for larger systems with a lesser level of precision.
When it comes to developments, it is important to found in literature related to computer-assisted drug design, which includes silico approaches with biophysical data, toxicology clinical studies, and other topics that are crucial in drug design and discovery. It aids computer-based concepts, which provide new ideas to staff in investigating and bringing theories together experimentally, as well as bringing answers to all difficulties, including cost, that arise. As a result, many more issues exist, such as the role of water molecules in overcoming constraints, solvent effects, entropic effects, receptor flexibility, and so on. This is a massive task that will lead to massive discoveries and provide information on the use of computerized technologies in drug design.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Sharma A (2023) Biological Effects of Narcotics Drugs. J Proteomics Bioinform. 16:647.
Received: 18-Jan-2023, Manuscript No. JPB-23-21447; Editor assigned: 20-Jan-2023, Pre QC No. JPB-23-21447 (PQ); Reviewed: 03-Feb-2023, QC No. JPB-23-21447;; Revised: 20-Apr-2023, Manuscript No. JPB-23-21447 (R); Published: 27-Apr-2023 , DOI: 10.35248/0974-276X.23.16.647
Copyright: © 2023 Sharma A. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.