Internal Medicine
Open Access
ISSN: 2165-8048
ISSN: 2165-8048
Review Article - (2020)Volume 10, Issue 3
Introduction: Aptamers are small single-stranded oligonucleotides (DNAs or RNAs) of low Mw that can form a specific 3d structure and can be produced by a sofesticated method known as Systematic Evolution of Ligands by EXponential enrichment (SELEX). aptamer technology has been used for medical and clinical purposes Due to their specific physical, chemical and biological properties.
Aim of the work: to study the aptamers from stuctural biology and its application in diagnosis and treatment modalities in different immuno-hematological disorders as aprospective solution.
Conclusions: We can safely conclude that aptamers are considered as a promising class of diagnostic and therapeutic agents.
Aptamers; SELEX; Immunohematology
Aptamers are small single-stranded oligonucleotides with low MW (ranges from 5-40 kDa), which form a specific 3d structure and can be produced by a hard technique known as Systematic Evolution of Ligands by Exponential enrichment (SELEX) which is based on repeated cycles of binding, separation, and amplification of oligonucleotides from a group of different sequences [1].
The reports have described aptamers targeted to a great variety of targets varying in nature and size ranging from ions to the whole cells, including small molecules, proteins, and viruses and peptides [1].
aptamers are defined as chemical antibodies and can be utilized for therapeutic applications [2].
aptamer has been used for medical and clinical applications. This review explains the uses of aptamers as a new tool for immunohematological applications [3].
Aptamers are short single stranded oligonucleotides ranges (~30 to ~70 nucleotides in length). In 1990 Ellington coined the term ‘aptamer’, derived from the combination of the Latin word aptus (to fit) and the Greek word meros (part).They fold into 3d structures and bind to their targets with high affinity and specificity and have a fixed sequence for binding the primer at both terminal ends for amplification using Polymerase Chain Reaction (PCR) [4].
Isolation of aptamers was made accessible due to the development of (SELEX). In this method, repeated cycles of selection, separation and amplification. Ligands of nucleotides were used until they show the greatest affinity and specificity to the target. The SELEX process starts with a large random pool of 1013-1015 oligonucleotides, which are chemically synthesized as DNA libraries (Figure 1) [5].
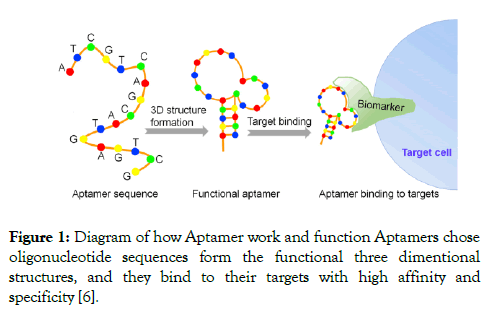
Figure 1: Diagram of how Aptamer work and function Aptamers chose oligonucleotide sequences form the functional three dimentional structures, and they bind to their targets with high affinity and specificity [6].
Aptamer vs Antibody
Aptamers in clinical application is gaining an advantages over antibodies, as discussed below:
1. Stability: proteins are denatured and lose the tertiary structure easily at elevated temperatures, while oligonucleotides are stable in high temperature and maintain their structures over iterative rounds of denaturation/Renaturation [7].
2. Synthesis: getting of monoclonal antibodies is produced inside labors and highly expensive procedure involving screening lots of colonies. However aptamers can be processed in quantity with high accuracy and specificity via chemical reactions [8].
3. Low immunogenicity: Aptamers characterized to be lessimmunogenic and less-toxic, because nucleic acids are small molecules and not recognized by the immune system. However, antibodies are highly immunogenic [9].
DNA and RNA aptamers
DNA and RNA aptamers are functionally similar but differ in their accessibility and stability. Compared with DNA aptamers, RNA aptamers are less stable than DNA aptamers due to the presence of a reactive hydroxyl group (-OH) in the 2 position of the ribose sugar in the RNA nucleotides. The nuclease resistance of RNA aptamers was found to increase when the 2`- hydroxyl group was removed from the sugars of RNA [10]. Because of the C-H bonds of the DNA nucleotides at position 2 of the deoxyribose sugar, DNA aptamers are less reactive and relatively stable [11]. So some modification chemically are usually done for RNA aptamers to increase their stability [12].
Strategies for chemical modifications of nucleic acid aptamers for improving the nuclease resistance of DNA aptamers, one of the functional solutions is Modifications of the sugar phosphate backbone or the pyrimidine are the strategies used to improve the stability and nuclease resistance of aptamers, capping the end of aptamers is one of the solutions for aptamer stability against nucleases, another approach to optain aptamer stability is Incorporation of unnatural nucleotides [13,14]. Locked nucleic acid (LNA) is one of the most prominent and successful nucleic acid analogues because of their pronounced stability, conjugation of polyethylene glycol (PEG) with DNA aptamer is commonly used to prolong their circulation in the bloodstream (Figure 2) [15,16]
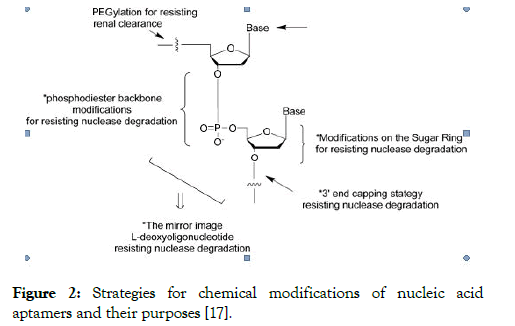
Figure 2: Strategies for chemical modifications of nucleic acid aptamers and their purposes [17].
Systematic evolution of ligands by exponential enrichment (SELEX)
Aptamers are selected from apool of an initial random singlestranded DNA depended on binding the oligonucleotides to the target molecules as (ions, organic dyes, amino acids, peptides, antibiotics, proteins, bacteria, viruses and even whole cells) under optimal conditions. The single-stranded DNA pool contains 1014~1016 single-stranded random oligonucleotides (about 40~100 nucleotides in the typical length, bounded by two constant regions with primer sites for polymerase chain reaction [PCR] amplification). The unbound sequences are isolated from the bound molecules, and the target-bound sequences are amplified by PCR (Figure 3) [18].
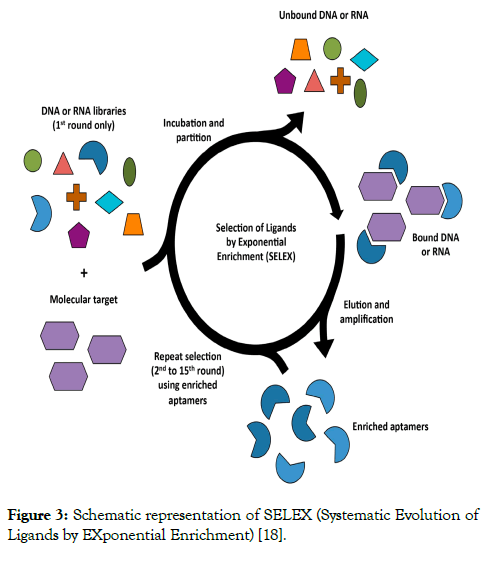
Figure 3: Schematic representation of SELEX (Systematic Evolution of Ligands by EXponential Enrichment) [18].
Aptamers as therapeutics
Aptamers as antagonists: Aptamers can inhibit enzyme activities or protein-protein interactions interactions after binding to the target.A number of aptamers are targeted against serum proteins, which are used in coagulation. Due to antagonistic properties, they have the potential to be used as anticoagulants to treat cardiovascular diseases and to prevent formation of blood clots during surgical interventions [19].
Aptamers as agonists: The agonistic aptamers can bind to the functional therapeutic relevant cell surface proteins and lead to an activation. The costimulatory receptor 4-1BB on the surface of activated T cells is a member of the tumor necrosis factor (TNF) receptor family and promotes the expansion and survival of activated T cells. McNamara and colleagues isolated an aptamer that targets and binds to 4-1BB expressed on activated mouse T cells surface [20].
Application of aptamers in different diseases
Apamers targeting leukemia - Diagnosis of leukemia biomarker: A panel of aptamers, called sgc aptamers was developed by Shuangguan and coworkers, for recognition of leukemia cell (Tcell lines, CCRF-CEM, human acute lymphoblastic leukemia) in the presence of normal bone marrow aspirates of ahuman. sgc8 aptamer (88-mer) showed the greatest affinity to its target [21].Later, sgc8c aptamer which is atruncated form of sgc8 aptamer (41-mer), was introduced with the same affinity and function.The target site of sgc8 aptamer on the surface of CCRF-CEM was discovered to be protein tyrosine kinase7 (PTK7) By using mass spectrometry (MS) and two-dimensional gel electrophoresis (2D-GE) [22].
Sefah et al introduced two aptamers called KH1C12 and KHG11 which could bind to the target AML cell line (Human promyelocytic leukemia cells, HL60) with high selectivity [23].
Nianxi Zhao’s group used hybrid SELEX approach to identify a recent ssDNA aptamer targets the therapeutic biomarker CD117, a transmembrane receptor expressed on AML cell [24]. The aptamer could be specifically and selectively bound to CD117 with great affinity, and was efficiently able to get internalized into CD117- expressing cells [21].
Aptamer-based targeted delivery system for treatment of leukemia: For acute myeloid leukemia (AML), an aptamer called As1411 was developed to be therapeutically used to target the membrane protein nucleolin which is a member of guanine-rich oligonucleotides (GRO) having antiproliferative and antitumor activity and highly expressed in cytoplasm and on the cell surface of many tumor types. It is being tested in phase II clinical trials for AML [25]. This protein in used in growth, proliferation, survival, nuclear transport, and transcription of the cells So, Inhibition of this protein by AS1411 aptamer affects several signaling pathways, such as Bcl-2 and NF-k-B [26]. It has been shown that AS1411 could effectively inhibit cell growth in MV4-11 after 72 h incubation (IC50=4 mM) [27].
Aptamers targeting immune system
Toll-like receptors: Toll-like receptors (TLRs) are a class of membrane spanning proteins that play important role in immune responses against invading microorganisms. TLRs can activate several signaling pathways such as mitogen-activated protein kinase (MAPK) and nuclear factor-kB (NF-kB) pathways to regulate immune response. Several aptamers to TLR have been designed. One of them Aptamer BL-7040 could bind to TLR9 and decrease AChE activity, which is thought to be beneficial in treating Sjogren’s syndrome or other autoimmune syndromes [28].
Cytokines: Cytokines are released by neutrophils, monocytes, macrophages as well as B cells and T cells. Several families of cytokine such as interleukins (IL), chemokines, interferons (INF), tumor necrosis family (TNF), and transforming growth factor (TGF) bind corresponding cell surface receptors and activate downstream signaling pathways to modulate immune responses [29]. Aptamers that bind cytokines have been developed to inhibit or promote the action of cytokines.IL-8 induces hemotaxis and phagocytosis for neutrophils and other granulocytes to migrate toward the site of infection. 8A-35 is an agonistic aptamer with neutralizing IL-8 activity and has potential to be effective therapeutic agent against inflammatory diseases [30].
Aptamers applications in multiple myeloma
One of the hopeful target for therapeutic treatment in multiple myeloma is CD38 which is a type II transmembrane glycoprotein and it is widely expressed on plasma cells and known to be important biomarker in MM [31].
Specific single stranded DNA aptamer targeted CD38 was produced capable of targeting multiple myeloma cells with great affinity then conjugation of this aptamer with Dox to form Apt- Dox conjugates (ApDCs); they found that there is specific internalization of the conjugated aptamer with Dox in multiple myeloma cells and there was continuous trafficking into lysosomes, where the low PH environment leaded to release of Dox from its conjugation to kill multiple myeloma cells without any harmfull to other normal cells [32].
Aptamers application therapy for SLE
RNA aptamer with a greater binding affinity to G6-9 which is an anti-DNA autoantibody and was produced by the MRL-lpr/lpr mouse model of human Systemic lupus erythromatosis (SLE) has been generated and is considered as an ideal target for the development of aptamers against SLE [33].
CCL2 is a small cytokine responsible for recruitment monocytes, memory T cells, and dendritic cells to the sites of inflammation produced by either tissue injury or infection and have a prominant role in human lupus nephritis of SLE [34].CCL2-specific binding aptamer called the spiegelmer mNOX-E36 could improve lupus nephritis and survival of MRLlpr/lpr mice by blocking the migration of monocytes from the bone marrow to sites of inflammation and decreasing the infiltrates of renal macrophage and T cell. Importantly, this aptamer did not have any immune-stimulatory adverse effects [35].
Aptamers for anticoagulation
The Archemix Corporation improved DNA aptamer called ARC-1779 to target the human von will brand factor (vWF) that interacts with GP1b on platelets and affects the P2Y12 receptor leading to platelet activation. The DNA aptamer ARC-1779 prevent platelet activation, platelet aggregation and thrombin generation. ARC-1779 was used in investigation of thrombotic thrombocytopenic purpura (TTP) [36]. A new anti-VWF RNA aptamer called DTRI-031 was produced recently. DTRI-031 was clinically used to prevent platelet aggregation in whole blood and inhibit thrombosis in mice models in a dose-dependent form [37].
Aptamers for hemophilia
BAX 499 (formerly ARC19499, Baxter Healthcare, Cambridge MA, USA) is an oligonucleotide aptamer developed in preclinical and clinical studies against Tissue factor pathway inhibitor (TFPI) which showed potent and specifc inhibition. In vitro studies demonstrated procoagulant activity with effective inhibition of TFPI, restoration of thrombin generation and clot formation in haemophilic plasma [38].
Aptamers are a special class of substances that combine the advantages typical both of low-molecular-weight substances and proteins, however the SELEX process is labour intensive, timeconsuming, and of relatively low yield (only about 30% of selections lead to the desired aptamers). Several variables need to be controlled carefully during SELEX, as they can influence the tertiary structure of aptamers. Some key variables include the incubation time, the ratio of aptamer to target, temperature, the size of the randomized library and the presence of monovalent and divalent cations. Optimization of these variables to control the yield and tertiary structure of these small aptamer molecules is even more challenging. These problems need to be addressed to enhance the diversity of aptamer libraries for selection of desired targets [39].
Modifications of nucleic acid aptamers with sugar moieties, phosphate linkages and nitrogenous bases, as well as multivalent modification using polymers such as polyethylene glycol (i.e. PEGylation) used to improve the nuclease resistance of DNA aptamers and Generation of highly specific aptamers with a long lifetime is very important for their therapeutic application [40].
Due to their unique physical, chemical, and biological properties.aptamer technology has been widely used in diagnosis and treatment of biomedical and clinical applications of immunohematology [3].
Although there has been great progress in aptamer and nanomedicine technology development in recent years, other challenges to these technologies should also be considered for examples: i) how can aptamers be modified in order to maintain the correct conformational structure on the surface of NPs? ii) How many aptamers should be conjugated with each NP to achieve effective binding with target cells? iii) How will the size and shape of the selected nanomaterials affect the nanomedicine biodistribution? iv) can the preparation method be easily and cost-effectively scaled up for industrial production?
Aptamers are attractive multifunctional tools that can be adapted for the development of nanomedicines. However, some challenges will have to be overcome in order to gain useful preclinical and clinical data, which will then lead to the translation of sophisticated, highly effective, aptamer-based nanomedicines from bench to bedside.
The authors declare no conflicts of interest regarding the publication of this paper.
Citation: Shoeib SA, Elhamid AEA, Zahran ES, Elkalashy ABE (2020) Aptamers from Biology to Find Solutions in Immunohematology10:315. doi: 10.35248/2165-8048.20.10.315.
Received: 19-Apr-2020 Accepted: 20-May-2020 Published: 27-May-2020 , DOI: 10.35248/2165-8048.20.10.315
Copyright: © Shoeib SA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited