
Immunotherapy: Open Access
Open Access
ISSN: 2471-9552

ISSN: 2471-9552
Research Article - (2022)Volume 8, Issue 4
Background: MMPs are a group of family genes that related to cancer progression, up regulation of most of the MMP genes in cancers are reported to be associated with promotion of invasion, angiogenesis, and immune surveillance avoidance. However, expression patterns of all MMPs in transcriptome level and single-cell level in a pan-cancer perspective have not been investigated.
Methods: Both the Cancer Genome Atlas (TCGA) transcriptome and single-cell sequencing pan-cancer data from GEO (Gene Expression Omnibus) were applied. The MMP-based diagnostic model was constructed by LASSO regression analysis. Tumors were classified into MMP score-high and low groups by ssGSEA. Single-cell data was analysisd by Serat package. The expression characters of MMPs were validated by qRT-PCR.
Results: MMP1, MMP11, and MMP12 were up regulated across almost all cancers. MMP19 and MMP27 are significantly down-regulated in eight to nine cancer types. Correlation analysis proved a potential relation between MMP expression and tumor immune and tumor stemness. Immune cells including Macrophages, Type II IFN Reponse, and Treg were highly infiltrated in MMP score-low group as expected, functions including Hippo signaling pathway, positive regulation of leukocyte adhesion to vascular endothelial cell, T cell chemo taxis more active in MMP score-high group. Single-cell analysis revealed diverse MMP expressions pattern in different cell clusters. In which, MMP7 are found to be highly expressed in the macrophages and MMP2 are found to be highly expressed in the CD8+T cells.
Conclusion: Majority of MMPs has increased expression across cancers, MMPs showed potential diagnostic value in combination.
MMP; Pan-cancer; Single-cell RNA sequencing; Tumor immune; Tumor stemness
Matrix metallopeptidases, a group of zinc-dependent endopeptidases, have been extensively investigated for their roles in pathways such as apoptosis, immunity, cellular migration, and angiogenesis by degrading the extracellular matrix [1]. To be specific, MMPs are divided into six subtypes according to different substrates: 1. collagenases, their main function is to degrade collagen I, II, and III: including MMP1 (collagenase-1), MMP8 (collagenase-2), MMP13 (collagenase-3); 2. gelatinases, their function is to degrade collagen I and IV: including MMP2 (gelatinase A) and MMP9 (gelatinase B); 3. Stromelysins: including MMP3 (stromelysin-1) and MMP11 (stromelysin-3); 4. MMP7. It is vital in the epithelization process of mucosal wounds, their substrates are fibronectin, laminin, and elastin; 5. MMP12, its substrates mainly include type IV collagen, gelatin, and fibronectin; 6. Membrane-type MMPs including MMP14, MMP15, MMP16, MMP17, MMP24, and MMP25. The substrates of MMP14 are type I, II, and III collagen. Their functions include keratinocyte growth, cell migration, and airway re-epithelization [2].
Meanwhile, many studies have found that MMPs help increase the invasion and metastasis of tumor cells [3,4], MMPs are also reported to be assistant of tumor cells to escape from the monitoring of the immune system, suggesting a significant role of MMPs play in carcinogenesis and tumor development [5,6]. In recent years, immunotherapy including cytokine treatment, cellular therapy, immune checkpoint blockades and many other methods has been proved to be successful in treating many fatal cancers [7,8]. This revealed the trend of finding new outcomes for cancer patients from the perspective of immunity. Recent studies have shown that MMP is highly associated with the microenvironment of cancers and immune cells, and targeted matrix metalloproteinases can overcome the obstacles of immuno-suppression [3]. Current researches mostly mention that continuous and automatic renewal of cancer stem cells endows cancer tissues with therapeutic resistance and relapse, which leads to worse prognosis and survival of cancers [9]. The relationship between the stem characteristics of cancer cells and MMP is still less discussed.
In this study, we systematically explored the relationship between expression levels of MMPs genomic changes in tumor tissues and cellular immunity in 18 cancers from the Cancer Genome Atlas (TCGA). On this basis, we further characterized the common points of MMPs expression at different immune levels from the single cell level. This article will explore the relationship between MMP in tumor tissues and stem characteristics. Our study has caused more thinking for the influence of MMP in the occurrence and development of cancer.
Pan-cancer transcriptome data and clinical data from TCGA, single-cell sequence data from GEO
mRNA expression and according clinical data, including tumor stage, age, gender and overall survival times of 18 types of cancer were downloaded from the Cancer Genome Atlas (TCGA) data portal [10]. The single-cell mRNA expression data was downloaded from NCBI GEO dataset (https://www.ncbi.nlm.nih.gov/gds) including hepatocellular carcinoma (GEO accession number: GSE103866), breast cancer (GSE123837), Colorectal Cancer (GSE97693), HNSC (GSE103322) and melanoma (GSE72056).
Transcriptome analysis
All statistical analyses and visualization of transcriptome data were performed by the R project (R version 3.6.1; R Foundation for Statistical Computing; www.r-project.org). Expression differences between cancer tissues and adjacent normal samples were examined by applying a t-test with the amount and significance of change. Survival data was obtained from the TCGA patient phenotype files. Kaplan-Meier survival analysis was used to compare survival differences in different groups.
Classification of MMP expressions activities across different cancer types
We selected an 18-gene expression signature (MMP1, MMP2, MMP3, MMP7, MMP9, MMP10, MMP11, MMP12, MMP14, MMP15, MMP16, MMP17, MMP19, MMP23A, MMP24, MMP26, MMP27, MMP28) according to the gene expression, their importance in cancer, and the previous study [11]. To classify MMP status, we employed GSVA [12] to calculate the MMP score based on the 18-gene expression signatures. The MMP score was calculated across all 18 cancer samples to classify samples as MMP score-high and MMP-low groups using top and bottom score samples.
Identification of pathways and clinical relevance analysis of MMP subtypes
We evaluated the associations of MMP score with patients’ overall survival time by using the R package “survival” and produce Kaplan-Meier survival plots, then, overall survival was evaluated by using the least absolute shrinkage and selection operator (LASSO) [13]. The difference of gene expression between MMP high and low groups was analyzed by edgeR, then, gene annotation and functional enrichment analysis of these genes were performed.
Single-cell data processing
Seurat R package were used for the preprocessing and the following analysis of the single-cell expression values. Cells that had RNA counts fewer than 200 or over 6000 were excluded from the downstream analysis, mitochondrial gene expression were also used as index for quality control. The remaining 5723 genes in 21621 cells passed quality control for the subsequent analysis. SC transform function of Seurat was used for data normalize. Principle component analysis (PCA) were used to analyze highly variable genes, the dimension reduction was conducted by using RunPCA function. Find Neightbor in Seurat were then used to process the significant PCA. Find Allmarker with resolution=0.5 was applied to identify the marker genes of each cluster, subsequently, each cell type of the cluster was defined by comparing gene markers to Cell marker database (http://biocc.hrbmu.edu.cn/CellMarker/index.jsp), including cancer cells, multi types of immune cells and multi types stromal cells. t-SNE method in the Rtsne package was used for visualization.
Patients
We collected 10 BC samples and corresponding para cancerous samples from IBC mastectomy patients for from October 2019 to December 2020 in the Endocrine and Breast Surgery department of the First Affiliated Hospital of Chongqing Medical University. All patients were diagnosed with molecular classification of triple-negative BC certificated by two pathologists in our department and all patients were provided with written informed consent. Samples were transferred in liquid nitrogen to store for further assessments. Original clinical data were collected from pathology reports and hospital records.
QRT-PCR
RNA was isolated from human tissues by applying the UNIQ-10 column RNA Extraction Kit (Sangon Biotech, China). Then, Reverse transcription was performed by the RR047 cDNA synthesis kit (TaKaRa, China). QRT-PCR was conducted by using 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA) with the 2 × Power SYBR® Green PCR Master Mix (Invitrogen, USA). Expression levels of gene were normalized to internal reference GAPDH. The sequences of primers are listed as follows:
MMP1-F (5′-atgtggagtgcctgatgtgg-3′),
MMP1-R (5′-ggctggacaggattttggga-3′),
MMP11-F (5′-cgacagaagaggttcgtgct-3′),
MMP11-R (5′-tccagcggtgcaatctcatt-3′),
MMP12-F (5′-tttggtggtttttgcccgtg-3′),
MMP12-R (5′-gcagcttcaatgccagatgg-3′),
MMP19-F (5′-tcaggtcagctggatgatgc-3′),
MMP19-R (5′-tcactcccatttgtccaggc-3′),
MMP24-F (5′-cagaaggtgaccccactgac-3′),
MMP24-R (5′-ccactgtgttgaagttgccg-3′),
GAPDH-F (5′-gcccgtttgcattttgtggag-3′), and
GAPDH-R (5′-ccaactttcgggaaatccat-3′).
MMP expression profiles in pan-cancer scale
Gene expression differences between tumor and normal tissues for the 18 MMP family members, including matrilysins, collagenases, stromelysins, and metalloelastase, were analyzed in 18 different cancer types by using the Cancer Genome Atlas (TCGA). Most MMP genes share similarities in expression patterns; they tend to up regulate in tumor tissue comparing with control tissue. Among them, MMP11, a stromelysin, shows the most significant dysregulation by significantly up regulating in most cancer types except KIRP and KICH (Figure 1). We have also noticed the up regulation of MMP1, MMP1, MMP19 and MMP12 in all tumor types, patients with lower expression of MMP11, and MMP12 also showed significant longer survival time (Figure 2). In the meantime, down regulation of MMP27, and MMP23B are found in various types of tumors, among them, MMP19 showed significant down regulation in tumor types of BLCA, BRCA, KICH, LUAD, LUSC, and UCEC, among them, LUSC patients and UCEC patients with lower expression of MMP19 showed a significant shorter survival time. Across 18 cancer types, most of them showed up regulation of MMPs, (eg: MMP1, MMP9, MMP10, MMP14, MMP15, MMP17, MMP11, MMP12), showed up regulation in BRCA, however Lung Squamous (LUSC) displayed distinct expression character from the other cancer types with significantly down regulated MMPs including MMP2, MMP15, MMP19, MMP23A, MMP24, MMP28. Additionally, UCEC showed lacked significant up regulation of MMPs comparatively (Figure 1).
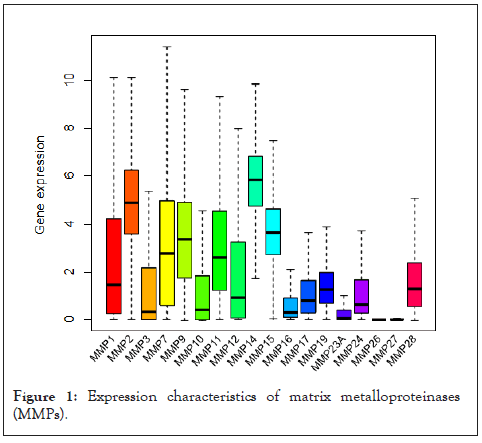
Figure 1: Expression characteristics of matrix metalloproteinases (MMPs).
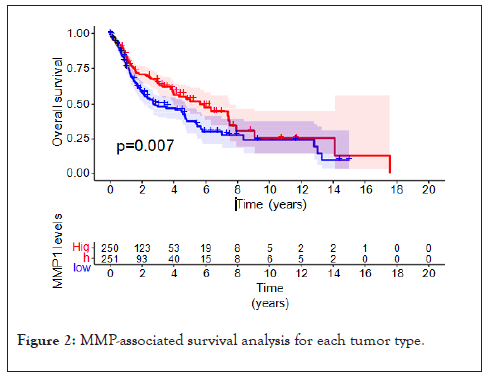
Figure 2: MMP-associated survival analysis for each tumor type.
Stemness and immune relation analysis of MMPs
The potential relativity between MMP expressions and tumor stemness and tumor immune were evaluated by applying the immune-score and the gene expression-based stemness index. The DNA stemness score (DNAss) had a significant positive correlation with MMP15 in most tumors and had negative correlation with most MMPs in TGCT. For the RNA stemness score (RNAss) there was a significant positive correlation with MMP2, MMP14, and MMP19, to be noted, MMP24 is also positively related with RNAss in LGG and TGCT. In addition [14], according to their study, the tumor microenvironment of all tumor was classified into six subtypes, including I: wound healing, II: IFN-γdominant, III: inflammatory, IV: lymphocyte depleted, V: immunologically quiet, and VI: TGF-βdominant. Our result showed that all hub genes were found to be expressed at higher levels in the VI subtype, which is also known for having the poorest prognosis. However, MMP1 showed a relative high expression in type III rather than type VI. Finally, we applied Cox regression to determine if MMP expressions were associated with patient survival. The forest plot results shows that the MMPs had a negative prognostic influence on most cancers, especially for MMP27, however, most MMPs hardly shows negative prognostic influence on SKCM (Figure 3).
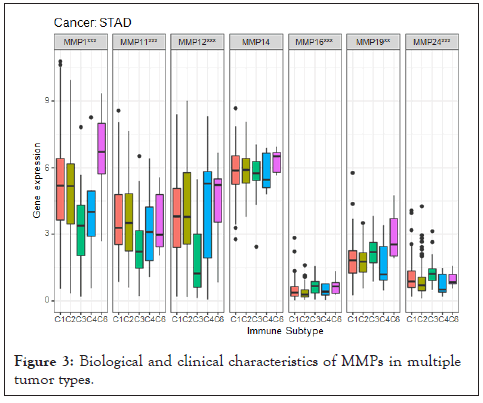
Figure 3: Biological and clinical characteristics of MMPs in multiple tumor types.
MMPs-based signature construction and TME phenotypes stratify
To examine whether tumors with the high and low MMP expressions were clinically distinct, we next construct risk score model according to the integrated data of MMP expression profiles and clinical characteristics in breast cancer. The LASSO analyses for the entire set revealed that MMP16, MMP26, and MMP27 were the top 3 most significant differently expressed MMPs. The prognosis scores were calculated as follows: risk score=0.017924 × expression of MMP1+0.035543 × expression of MMP3+0.003835 × expression of MMP7+0.011903 × expression of MMP9+0.002472 × expression of MMP11+0.015078 × expression of MMP15+0.028677 × expression of MMP16+0.003561 × expression of MMP17+0.021418 × expression of MMP19+0.023968 × expression of MMP24+0.117175 × expression of MMP26+0.143249 × expression of MMP27. The expression of the most above genes was negatively related to the overall survival of breast cancer patients. Based on the median risk score, patients were divided into high-risk and low-risk groups. The survival probability of the low-risk group was significantly higher than that of high-risk group individuals. To further detect the overall biological function of the MMP family in tumors, we used MMP expression based-ss GSEA score to classify the tumor samples. As shown in Figure, tumors were divided into MMP score-high and low groups, differential signature enrichment were conducted by applying GSVA analysis, The result showed that immune cells including Macrophages, Type II IFN Reponse, and Treg were highly infiltrated in MMP score-low group as expected, functions including Hippo signaling pathway, positive regulation of leukocyte adhesion to vascular endothelial cell, T cell chemotaxis more active in MMP score-high group (Figure 4).
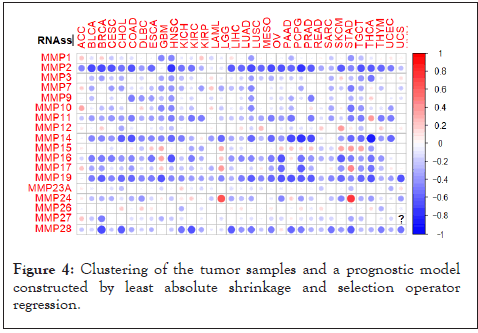
Figure 4: Clustering of the tumor samples and a prognostic model constructed by least absolute shrinkage and selection operator regression.
Landscape of MMP expressions at single-cell level
To determine the single-cell level transcriptomic pan-cancer landscape, we applied scRNA-sequencing data from multiple types of tumors including melanoma, head and neck squamous cell cancer, breast, liver, and colon cancer. We first performed quality control to the gene expression matrix to screen genes and cells. Next, normalization of scRNA-seq data was conducted and we screened 20 principal components for subsequent analysis (Figure 5). Then we used t-distributed stochastic neighbor embedding (t-SNE) method for the unsupervised analysis of cell clustering. The result revealed high cell heterogeneity of tumor tissue, in which tumor cells were segregated into 16 distinct clusters, including multiple types of tumor cells (including cluster 0, 1, 3, 10, 12, 14, 15), immune cells: Including macrophages (cluster 2), CD8+T cells (cluster 5,8), CD4+T cells (cluster 11), B cells (cluster 16), and NK cells (cluster 9), non-immune cells: including tumor-associated fibroblasts (cluster 4, 7), endothelial (cluster 6), and other unidentified cells (cluster 13), which were defined by single R and cell markers. Next, we tested the expression pattern of the MMPs in these cell clusters. Most of the MMPs were highly expressed in tumor cells as expected, to be noted, MMP7 are found to be highly expressed in the macrophages and MMP2 are found to be highly expressed in the CD8+T cells (Figure 6).
Figure 5: Pre-processing of the single-cell sequencing data and cell cluster identification.
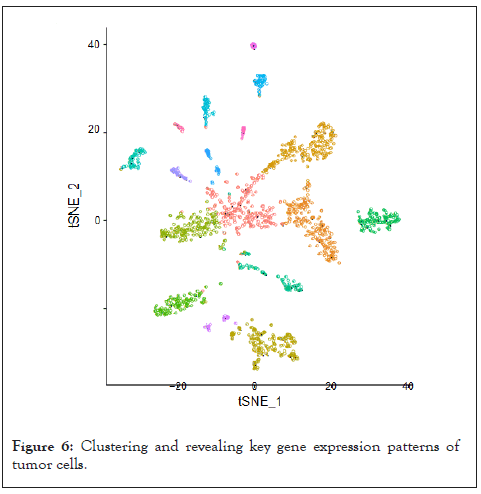
Figure 6: Clustering and revealing key gene expression patterns of tumor cells.
To further uncover the detailed heterogeneity of tumor cells, we took a better look at the eight cell clusters of the tumor cells, including clusters 0, 1, 3, 10, 12, 14, and 15, Based on the gene markers, clusters 0, 1 and 3 were defined as immune-related cell groups, 8 and 10 were defined as metabolic-related cells, cluster 12, 14, 15 were defined as extracellular matrix-related cells.
Differential expression of MMPs in tumors and adjacent tissue
The expression levels of MMP1, MMP11, MMP19, and MMP24 in different tissues were detected by qRT-PCR. It was found that the expression of MMP1, MMP11, MMP12 in tumor tissue were dramatically higher than that in adjacent breast tissue (10 cases) and the expression of MMP19 and MMP24 in tumor tissue were significantly lower than that in adjacent breast tissue (Figure 7).
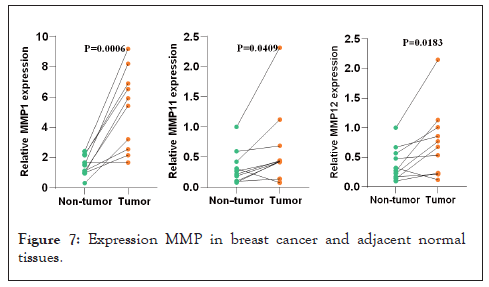
Figure 7: Expression MMP in breast cancer and adjacent normal tissues.
Former studies have indicated that compared with normal tissues, the expression levels of various MMPs in tumor tissues show significant differences [15]. Our study discovered the differential expression of 18 MMP members in 18 cancer types, where MMPs were more commonly up-regulated in tumor tissue than in normal tissue, which consistent with the conclusion of previous studies. At the same time, our analysis also verified that MMP-2 and MMP-9 were the most commonly deregulated proteases in all MMP families [16], which further supported the reliability of our study. In addition, we supplemented the previous description of MMP expression in pan-cancer. Among them, MMP-1, an interstitial collagenase, was up-regulated in 15 of 18 types of tumors. For example, in HNSC, the expression was up-regulated nearly 7.5-fold as compared to the control group. Meanwhile, the expression of MMP-1 in LUSC and BRCA was also significantly increased.
Head and Neck Cancer (HNSC), the seventh most common cancer in the world, accounts for 3% of all cancers and more than 1.5% of cancer deaths in the United States [17]. We discovered significantly changed of various MMP expression levels in head and neck cancer as compared with normal tissues which are accord with the previous study reported that MMP-1, 3, 7, 8, 10, 12, 20 and 27 genes showed multiple mutations in patients with head and neck cancer, they also confirmed a positive correlation between MMP-20 expression levels and the staging of the cancer [18]. Among these, MMP-1 is considered to be the most potential marker of HNSC, and it can promote disease progression by activating AKT pathway [19]. An association between MMP-13 expression and the younger diagnostic age of HNSC [20]. MMP-17 has also been found to strengthen the metastasis of head and neck cancer cells in a hypoxia environment through promoting the invadopodia formation and amoeboid movement [18].
As a tumor with significantly different MMP gene expression in this study, the complex entanglement between lung cancer and MMPs has been studied more often. We found that the expression levels of MMPs in different histological types of lung cancer were not in step, which is consistent with the conclusions of previous studies. Increase of more than a 4-fold of MMP-1, 8, 9 and 12 expressions in squamous cells compared to adenocarcinoma [21]. A recent study observed that single-stranded RNA (sgRNA) targeting MMP-8 reduced the proliferation of human lung adenocarcinoma cells and reduced tumor mobility, using MMP-8 sgRNAs with CRISPR-Cas9 [22]. On this basis, the latest research has found that the down-regulation of MMP-20 gene in human lung adenocarcinoma cancer cause cells death and reduce cells migration by inhibiting the expression of PI3K and survivin genes [23]. Patients with lung cancer will stimulate the expression of P53, and this tumor suppressor protein gene is up-regulated through gene mutation, and its dysfunction stimulates the occurrence of tumors. The synergistic effect of that two reduce the three-year survival of patients with lung cancer from 28% to 14% (p=0.001) [24], which is consist with our study.
According to statistics, breast cancer has now surpassed lung cancer as the most common tumor in the world in 2020, and the pandemic of COVID-19 may cause more delay in diagnosis [25], which means that the number of breast cancer patients are underestimated, which has forced people to become more enthusiastic about breast cancer research. In relation to the MMPs family and breast cancer, the latent form expression of MMP-2 and MMP-9 appear to be related to breast cancer risks [26]. Ets-1, as an important transcription factor regulating invasion and metastasis of tumor, has its binding site on MMP-9 promoter [27], which explains the role of MMP-9 in promoting invasion of breast cancer. A recent study has found that MMP-1 induced brain metastasis of breast cancer is related to the down-regulation of non-coding RNAMiR-202-3p in brain metastases [28]. In addition to promoting distant metastasis of cancer cells, MMP1 also contributes to a worse survival for patients with Her2 and basal-like subtype breast cancers [29]. All these are consistent with the adverse effects on survival of high expression of MMP1 in our study. In the treatment of breast cancer, found in their experiment that serum levels of tissue inhibitor of MMP-1 and MMP-3 (TIMP-1, TIMP-3) were down-regulated with radiotherapy and inversely proportional to treatment time [30], which was a new idea for predicting radiotherapy toxicity. Another study confirmed that the combined reaction of MMP-14 and integrin β1 induced new DNA damage reactions (stalling of replication forks and double strand breaks) and increased the sensitivity of TNBC cells to ionizing radiation and doxorubicin by in vitro and in vivo breast cancer models [31], combined with our study, these provided new targets for the treatment of breast cancer.
As both the first and possibly the last line of defense, anti-tumor immunity profoundly influence the outcome of tumor by forming diverse types of inflammatory reactions in tumor microenvironment, while adaptive immunity plays an important role in the anti-tumor immunity. It is reported that the expression of MMP in immune inflammatory factors is triggered by multiple transcription factors on the gene promoter. For example, interleukin-12 (IL-12) can mediate the activation of different Signal Transducer and Activator of Transcription (STATs), resulting in enhanced transcription of interferon gamma (IFN-γ), and finally affecting the expression of MMP-2 and MMP-9. IL-12 could also activate the NF-κB (nuclear factor-κB) pathway and induce the expression of MMP-1 and other MMPs [32]. We found Macrophages, Type II IFN Reponses, and Treg were relatively high in MMP-score low group. MMP-13 in MMP family down-regulated the ability of mouse bone marrow-derived DCs to activate CD8+ T cells by reducing MHC-1 surface presentation, endocytosis, and cytokine/chemokine secretion [33]. MMP-11 expression in the mono nuclear infectious cells (MIC) of breast cancer was positively correlated with the ratio of tumor-frontier inflammatory factor CD68/(CD3+CD20) [34]. A recent study by Kim et al. also suggested that high MMP-11 expression was associated with genome-related down-regulation of CD8 T cells, CD4 T cells, and memory B cells, respectively [35].
Our single-cell analysis shows that MMP7 is highly expressed in macrophages, and MMP2 are found to be highly expressed in the CD8+T cells. Previous studies have found that lymphocytes mainly secrete MMP-2 and MMP-9, have shown that MMP-2 expression is particularly up-regulated in Th1 lymphocytes, which are more capable of promoting local ECM degradation than other phenotypes [36].The expression of MMP-9 was inversely proportional to the T cell diversity and the number of T helper cell type 1 cytokines in the tumor, and its down-regulation promoted the entry of effector/memory T cells into the tumor [37]. On this basis, a recent study reduced the mRNA and protein levels of PD-L1 in cancer cells through the inhibition of MMP-2 and MMP-9 by 7B-3CT [38], which inspired new MMPs targets for Immune Checkpoint Blockade (ICB) therapy. The differential expression of MMPs also offers a new direction for early diagnosis and detection of tumors. Previous studies proved the immune response of MMP-11 in the cytoplasm and fibrous matrix of breast cancer and it was also actively expressed in the blood flow of her patient and in the autoantibodies [39]. These results suggest that tumor associated immune cells are deeply involved in the MMP expression.
Our results showed that expressions of many MMPs are associated with stemness of tumor, Former study have noticed this potential connection by discovering interaction between MMPs and Dentin Sialo Phospho Protein (DSPP) in oral Squamous Cell Carcinoma (OSCS), it is also found that the double silencing of DSPP and MMP-20 gene resulted in the decrease of cancer stem cells (CSCs) expressions, which also increased the sensitivity of OCSC to drugs such as cisplatin [40]. We have also found that the up regulation of MMP expressions is related with Hippopotamus pathway. It has been reported that high-frequency mutations of Hippo pathway appear in human malignant mesothelioma and meningioma, and the study on the relaxation direction of this pathway has also inspired further exploration on the efficacy of Hippo-targeted therapy in solid malignant tumor [41]. To be noted, we are not the first one to be interested in the connection between the hippo pathway and MMP family, recent studies have found that while inhibiting the hippopotamus pathway, such as TNFAIP8 and SCC-S2, can affect the outcomes of cancers such as lung cancer and colorectal cancer, and are associated with the abnormal expression of MMP-7 in MMP family members.
This study is the largest MMP gene analysis to date for it encompasses 18 MMP genes by using RNA-seq and SCRNA-seq technology across 18 different types of cancers. Based on this manuscript, we have revealed diagnostic capabilities of the MMPs across cancers. Functional studies of these genes showed potential relations between MMP expressions, tumor immune, and tumor stemness.
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
Citation: Tan J, Gao Y, Li Z, Wang Y, Wang L (2022) A Perspective of Matrix Metalloproteases (MMP) Gene Expression Profile Revealed by Pan-cancer Transcriptome and Single Cell Data. Immunotherapy (Los Angel). 8:199.
Received: 29-Jun-2022, Manuscript No. IMT-22-18189; Editor assigned: 01-Jul-2022, Pre QC No. IMT-22-18189 (PQ); Reviewed: 15-Jul-2022, QC No. IMT-22-18189; Revised: 22-Jul-2022, Manuscript No. IMT-22-18189 (R); Published: 05-Aug-2022 , DOI: 10.35248/2471-9552.22.8.199
Copyright: © 2022 Tan J, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.