Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research Article - (2022)Volume 15, Issue 6
To invade human cells, COVID-19 uses its spike glycoprotein as the major surface protein. This glycoprotein can be inactivated through chemical modification of surface accessible asparagine and lysine residues by vitamin B6 (pyridoxal-5'-phosphate, PLP) and vitamin C (ascorbic acid). This reversible reaction leading to the formation of a Schiff base (aldimine), and consequent reduction by ascorbate lead to form a stable phosphopyridoxyl-lysyl or asparagyl covalent bonds. For this purpose, at first, the spike glycoprotein sequences in similar proteins with 100% identity was specified by using of UniProtKB that is located at the ExPASy server. All of these proteins have a 1,255 length and 139,125 Mass (Da) respectively. Then, GetArea software was used for accessible surface area determination of each sequence by employing of PDB files that is reserved at PDB website. The results revealed that, 51 out of 81 (for asparagine) and 22 out of 48 (for lysine) are accessible for PLP reaction. Thus, terminal amino groups (NH2) of asparagine and lysine can react with PLP and produce Schiff base bond an then it can be converts to a strong covalent bond by ascorbic acid reduction. In this way, attachment and fusion process can be prevented. This may be a way for vaccine and drug preparation.
Spike glycoproteins; COVID-19; Vitamin B6; Chemical modification; Bioinformatics
Severe Acute Respiratory Coronavirus 2 (SARS-CoV-2) was determined to be the agent of coronavirus disease 2019 (COVID-19). The virus belongs to the Betacoronavirus genus of the Coronaviridae family, which also includes Severe Acute Respiratory Syndrome Coronavirus 1 (SARS-CoV-1) and Middle East Respiratory Syndrome Coronavirus (MERS-CoV) [1]. The pandemic caused by the severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) has generated global concern given its rapid spread in multiple countries with fatal progression in a considerable proportion of COVID-19 patients [1,2]. SARS-CoV-2 is an enveloped virus with a positive-sense, single-stranded RNA genomewith sizes ranging from 26-32 kilobases im length [3]. The SARS-CoV-2 genome encodes four major structural glycoproteins to produce a structurally complete viral particle which includes the nucleocapsid, membrane, envelope, and spike proteins (S) [4]. This kind of particle can enter host cells by binding the viral surface spike glycoprotein to angiotensin-converting enzyme 2 [5]. The spike glycoproteins forms homo-trimers protruding form the viral surface; especially, the spike 1 surface unit allows the attachment of the virus to cellular receptors [4,5]. By using of several bioinformatics tools and biological databases functional analysis for prediction of spike glycoprotein inactivation can be possible. Bioinformatics study is a strong tool specified in sortation, organization, and process large amounts of available data generated from other experiments to provide a large-scale immunological platform within a limited time. Since the virus genome and its protein sequences information are available, the presented epitopes and the virus characteristics could be predicted by in silico analysis, which significantly accelerates the progress of vaccine development [6-10].
The SARS-CoV S glycoprotein is synthesized as a 1255 amino acid polyprotein precursor on the rough endoplasmic reticulum (RER) [11]. The unprocessed precursor harbors an endoplasmic reticulum (ER) signal sequence (chain 1-13) located at the N terminus, which targets the S glycoprotein to the RER membrane and is removed by cellular signal peptidases in the lumen of the ER [12,13]. In the trans-Golgi network, the SARS-CoV-2 S glycoprotein is proteolytically cleaved by cellular furin or furin-like proteases at the S1/S2 cleavage site, comprising multiple arginine residues that are not found in the closely related SARS-CoV [14,15]. Cleavage at the S1/S2 site yields a surface subunit S1, which attaches the virus to the host cell surface receptor, and a transmembrane subunit S2, which mediates the fusion of viral and host cell membranes [16]. The S1 and S2 subunits remain associated through noncovalent interactions in a metastable prefusion state [17]. Furin-like cleavage is essential for the S-protein mediated cell-cell fusion and viral infectivity, and is required for efficient SARS-CoV-2 infection of human lung cells [14] and airway epithelial cells [18]. Membrane fusion and viral entry of SARS-CoV-2 is initiated by binding of RBD in the viral S glycoprotein transiently sampling the functional conformation to ACE2 on the surface of target cells [19]. These spike glycoproteins are included; Spike protein S1, that, attaches the virion to the cell membrane by interacting with host receptor, initiating the infection (By similarity). Binding to human ACE2 and CLEC4M/DC-SIGNR receptors and internalization of the virus into the endosomes of the host cell induces conformational changes in the S glycoprotein. Proteolysis by cathepsin CTSL may unmask the fusion peptide of S2 and activate membranes fusion within endosomes. Spike protein S2 which, mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least three conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell. membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.UniRule annotation. Spike protein S2' that, acts as a viral fusion peptide which is unmasked following S2 cleavage occurring upon virus endocytosis. S1, S2, and S2' locations that can link by glycosylation via N-linked of asparagine (GlcNAc...) and host membrane cell (Figure 1) [19].
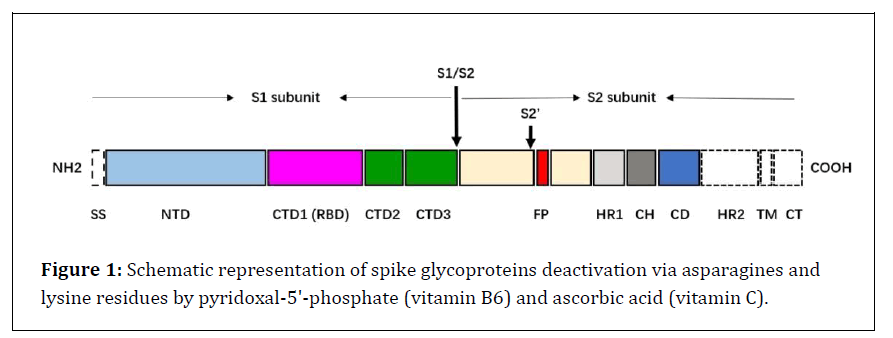
Figure 1: Schematic representation of spike glycoproteins deactivation via asparagines and lysine residues by pyridoxal-5'-phosphate (vitamin B6) and ascorbic acid (vitamin C).
This study is aimed at prediction of Spike SARS-CoV-2 glycoproteins inactivation by chemical modification of asparagine and lysine residues via covalently binding to pyridoxal-5'-phosphate (vitamin B6) and then reduction of Schiff base formed by vitamin C. Due to the binding of spike glycoproteins to ACE2 human cells, which is mostly done through asparagine, the main emphasis in this study is on asparagine roots (Figure 2).
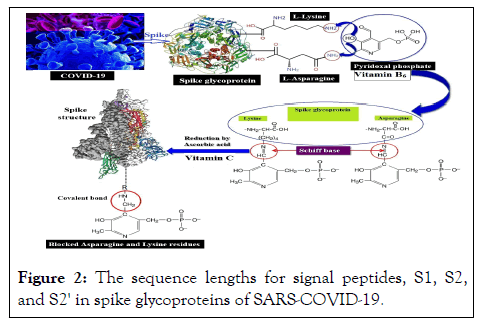
Figure 2: The sequence lengths for signal peptides, S1, S2, and S2' in spike glycoproteins of SARS-COVID-19.
The amino acid sequences of SARS-CoV spike glycoprotein and docking was gained from UniProtKB and Swiss-Dock that are located at the ExPASy server (Swiss Bioinformatics Resource Portal). The structure of glycoproteins as well as, pdb data files were resulted from The Protein Data Bank (PDB; http://www.rcsb.org/pdb/). Calculation of Solvent Accessible Surface Area was performed by GETAREA from sealy center for structural biology, university of Texas medical branch, Galveston. TX 77555. Docking of spike glycoprotein with pyridoxal-5'-phosphate (as a ligand) was carried out by using of SwissDock that is located at the ExPASy server.
Spike glycoprotein S1, S2, and S2' sequences
Based on information that is located at UniProtKB (ExPASy server) for Spike glycoprotein, P59594|SPIKE_CVHSA Spike glycoprotein OS=Human SARS coronavirus, the sequence lengths for signal peptides, S1, S2, and S2' are as follow in Figure 3.
Figure 3: Schematic representation of the domain arrangement of the SARS-CoV-2 S protein precursor. SS, signal peptide; NTD: N-terminal domain; RBD: receptor-binding domain; RBM: receptor-binding motif; SD1/2: subdomain 1 and 2; FP, fusion peptide; HR1, heptad repeat 1; CH, central helix; CD, connector domain; HR2, heptad repeat 2; TM, transmembrane domain; CT, cytoplasmic tail. Arrows denote protease cleavage sites [19].
ExPASy Code: P59594|SPIKE_CVHSA Spike glycoprotein OS=Human SARS coronavirus (Severe acute respiratory syndrome coronavirus) Length:1,255; Mass (Da):139,125; Total Asparagine residues (Asn, or N)=81(Table 1).
| Chains | Length (Amino acids) |
|---|---|
| Signal peptides, 1-13 | 13 |
| Spike glycoprotein, 14-1255 | 1242 |
| Spike protein S1 UniRule annotation, 14-667 | 654 |
| Spike protein S2 UniRule annotation, 668-1255 | 588 |
| Spike protein S2', 798-1255 | 458 |
Table 1: Spike glycoprotein sequences. Signal peptides, Spike protein S1, S2, and S2' chain and amino acid content.
Glycosylation regions by Asparagine at host cell membrane are present below:
Glycosylation 29 N-linked (GlcNAc...) asparagine
Glycosylation 65 N-linked (GlcNAc...) asparagine
Glycosylation 73 N-linked (GlcNAc...) asparagine
Glycosylation 109 N-linked (GlcNAc...) asparagine
Glycosylation 118 N-linked (GlcNAc...) asparagine
Glycosylation 119 N-linked (GlcNAc...) asparagine
Glycosylation 158 N-linked (GlcNAc...) asparagine
Glycosylation 227 N-linked (GlcNAc...) asparagine
Glycosylation 269 N-linked (GlcNAc...) asparagine
Glycosylation 318 N-linked (GlcNAc...) asparagine
Glycosylation 330 N-linked (GlcNAc...) asparagine
Glycosylation 357 N-linked (GlcNAc...) asparagine
Glycosylation 589 N-linked (GlcNAc...) asparagine
Glycosylation 602 N-linked (GlcNAc...) asparagine
Glycosylation 691 N-linked (GlcNAc...) asparagine
Glycosylation 699 N-linked (GlcNAc...) asparagine
Glycosylation 783 N-linked (GlcNAc...) asparagine
Glycosylation 1056 N-linked (GlcNAc...) asparagine
Glycosylation 1090 N-linked (GlcNAc...) asparagine
Glycosylation 1116 N-linked (GlcNAc...) asparagine
Glycosylation 1140 N-linked (GlcNAc...) asparagine
Glycosylation 1155 N-linked (GlcNAc...) asparagine
Glycosylation 1176 N-linked (GlcNAc...) asparagine
Solvent accessible surface area assessment by GETARE
For asparagine residues estimation that are located at the surface of spike glycoprotein and can be react with pyridoxal phosphate, the spike glycoprotein sequences were analysed by GETAREA software. For this reason, at first pdb format files of spike glycoproteins with pdb code 5WRG (method: Electron microscopy, Resolution: 4.30 Å, Uniprot: P59594) and 1WNC (method: X-Ray Diffraction, Resolution: 2.80 Å, Uniprot: P59594 ), that was placed in the PDB website were selected. Then, pdb format files were uploaded to GETAREA software with water Probe radius of 1.400 Å and the area/energy/residue was calculated. Afterwaeds, the total numbers of each amino acid, the number of amino acids available at the surface (out) and the number of amino acids within the spike glycoprotein molecule (In) were estimated (Table 2).
| Amino acid | Total number | Accessibility | |
|---|---|---|---|
| In | Out | ||
| THR | 78 | 45 | 33 |
| PHE | 42 | 37 | 5 |
| MET | 12 | 11 | 1 |
| LEU | 60 | 54 | 6 |
| LYS | 48 | 26 | 22 |
| TYR | 34 | 29 | 5 |
| ASP | 60 | 36 | 24 |
| GLU | 23 | 12 | 11 |
| ASN | 81 | 30 | 51 |
| GLY | 46 | 33 | 13 |
| ILE | 42 | 35 | 7 |
| VAL | 57 | 49 | 8 |
| ALA | 52 | 40 | 12 |
| CYS | 37 | 37 | 0 |
| SER | 59 | 42 | 17 |
| GLN | 38 | 28 | 10 |
| PRO | 35 | 24 | 11 |
| ARG | 26 | 23 | 3 |
| TRP | 4 | 3 | 1 |
| HIS | 6 | 4 | 2 |
Table 2: Accessible surface area results for all 1255 amino acids (pdb: 5WRG).
Spike glycoprotein docking with pyridoxal-5'-phosphate
Docking of spike glycoprotein (pdb code: 1WNC) with pyridoxal-5'-phosphate was achieved by using of SwissDock (Docking of small ligands into protein molecules) that is located at the ExPASy server (Swiss Bioinformatics Resource Portal).
Spike glycoprotein S1, S2, and S2' sequences
Based on sequence data for spike glycoprotein (Uniprot code: P59594) that is located at UniprotKB at ExPASy server, Protein: (Spike glycoprotein, Gene: S, Organism: Human SARS coronavirus (SARS-CoV) (Severe acute respiratory syndrome coronavirus), there are 81 asparagine amino acid residues in the COVID-19 glycoprotein sequence that are highly reactive with pyridoxal-5'-phosphate. These asparagines are marked in green in sequence (Figure 4).
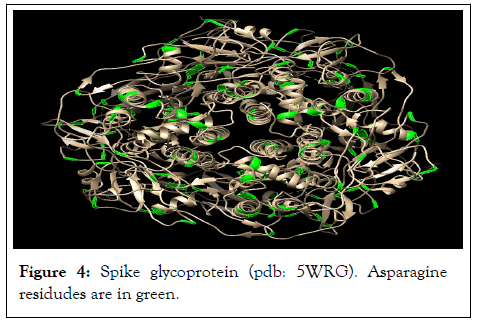
Figure 4: Spike glycoprotein (pdb: 5WRG). Asparagine residudes are in green.
Solvent accessible surface area assessment by GETARE
The results of GETAREA revealed that, among these 81 asparagine (Asn) residues that are present in the spike glycoprotein sequences, 51 Asn are at the surface of the protein molecule (Outer) and 30 Asn are within the protein structure. Hence, there is a highly chance for amino groups (NH2) of Asparagine (Asn) reaction with pyridoxal-5'-phosphate and reduction of these schiff base by ascorbic acid (vitamin C) for producing covalently bonded pyridoxal phosphate to Asn residues and inactivation of reactive region of spike glycoproteins or inhibition of Glycosylationi N-linked (GlcNAc...) asparagine with the host cells (Table 3).
| ASN Num |
Accessibility | ASN Num |
Accessibility | ASN Num |
Accessibility | ASN Num |
Accessibility |
|---|---|---|---|---|---|---|---|
| 29 | Out | 330 | Out | 699 | Out | 1107 | - |
| 65 | Out | 347 | Out | 721 | In | 1116 | Out |
| 73 | Out | 357 | Out | 733 | Out | 1140 | Out |
| 78 | Out | 375 | Out | 746 | In | 1155 | Out |
| 96 | Out | 381 | In | 759 | In | 1160 | Out |
| 108 | Out | 409 | In | 783 | In | 1169 | Out |
| 109 | Out | 424 | In | 806 | Out | 1174 | Out |
| 118 | Out | 427 | Out | 827 | In | 1176 | Out |
| 119 | Out | 435 | In | 838 | In | 1176 | Out |
| 122 | In | 437 | Out | 889 | Out | ||
| 129 | In | 457 | Out | 896 | Out | ||
| 135 | Out | 473 | Out | 901 | In | ||
| 155 | In | 479 | Out | 907 | Out | ||
| 158 | In | 505 | Out | 910 | Out | ||
| 178 | In | 522 | Out | 935 | In | ||
| 189 | In | 526 | In | 937 | In | ||
| 214 | In | 528 | In | 942 | Out | ||
| 227 | Out | 530 | In | 951 | In | ||
| 230 | Out | 589 | Out | 960 | In | ||
| 269 | Out | 602 | Out | 1005 | In | ||
| 281 | Out | 626 | Out | 1056 | Out | ||
| 304 | In | 627 | Out | 1080 | - | ||
| 318 | Out | 691 | Out | 1090 | Out | ||
| 321 | Out | 692 | Out | 1101 | Total ASN Number=81 residues | ||
Table 3: GETARE results for spike glycoprotein Asparagine residues.
Spike glycoprotein docking with pyridoxal-5'-phosphate
The results of spike glycoprotein docking (1WNC) with pyridoxal-5'-phosphate as a ligands confirmed the results of Accessible Surface Area evaluations and implied that, pyridoxal phosphate can bind to the spike glycoprotein via asparagine residues that are present at the outer surface of this protein molecule (Figure 5).
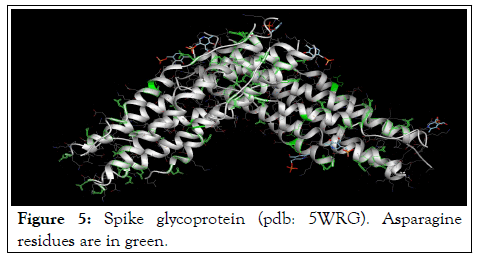
Figure 5: Spike glycoprotein (pdb: 5WRG). Asparagine residues are in green.
Realizing every step during virus entry into the human cell and of virus replication and packaging could, in principle, be the aim of inhibitors. Identification of asparagine residues in reaction with human cell ACE2 by using of bioinformatic tools for inactivation of these residues by chemical modification with pyridoxal phosphate (vitamin B6) and ascorbic acid (vitamin C) will provide the insights to assist in the development of SARS-CoV-19 vaccines.
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Google Scholar][PubMed].
[Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
Citation: Torabizadeh H (2022 A Bioinformatic Approach for COVID-19 Spike Glycoprotein Deactivation by Chemical Modification of Asparagine and Lysine Residues via Vitamin B6 and Vitamin C. J Proteomics Bioinform. 15:591.
Received: 14-Jun-2022, Manuscript No. JPB-22-17927; Editor assigned: 17-Jun-2022, Pre QC No. JPB-22-17927 (PQ); Reviewed: 01-Jul-2022, QC No. JPB-22-17927; Revised: 08-Jul-2022, Manuscript No. JPB-22-17927 (R); Published: 15-Jul-2022 , DOI: 10.35248/0974-276X.22.15.591
Copyright: © 2022 Torabizadeh H. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.