Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Mini Review - (2021)Volume 14, Issue 10
A Spatial and Single-cell Proteomics Workshop on how to map the location and connection of proteins was hosted virtually by the Plant Cell Atlas (PCA) initiative on April 8th, 2021. The objective of the PCA is to form a community to discuss novel developments in high-resolution subcellular and cellular localization, mass spectrometry and proteomic workflows in plant science. This workshop hosted six top scientists including three established investigators and three rising stars in the areas of spatial and quantitative proteomics from plant and animal sciences that contribute to the technological advances of modern proteomics. The goal of this workshop is to highlight current advancements in cellular and subcellular protein localization patterns, optimization of techniques in single-cell proteomics, and mapping of protein complexes and protein-protein interactions (PPI).
Spatial proteomics; Single-cell proteomics; Protein-protein interaction; TurboID
Proteins are important functional molecules in cells. Proteomics aims to identify, quantify, and characterize all the proteins in a cell, tissue, or organism at any given time and environmental condition. Mass spectrometry (MS) facilitates determination of protein properties, such as sequence, quantity, post-translational modifications, interacting partners, as well as subcellular localizations and trafficking.
In the past decade, MS-based proteomics has uncovered many disease biomarkers and improved understanding of cellular processes, molecular networks, dynamic molecular changes, and the biological functions. With major advances in spatial and single-cell proteomics, this workshop was designed to cover three fast-growing areas by six invited speakers: spatial and subcellular proteomics, single-cell proteomics, and protein complexes and protein-protein interactions (Figure 1).
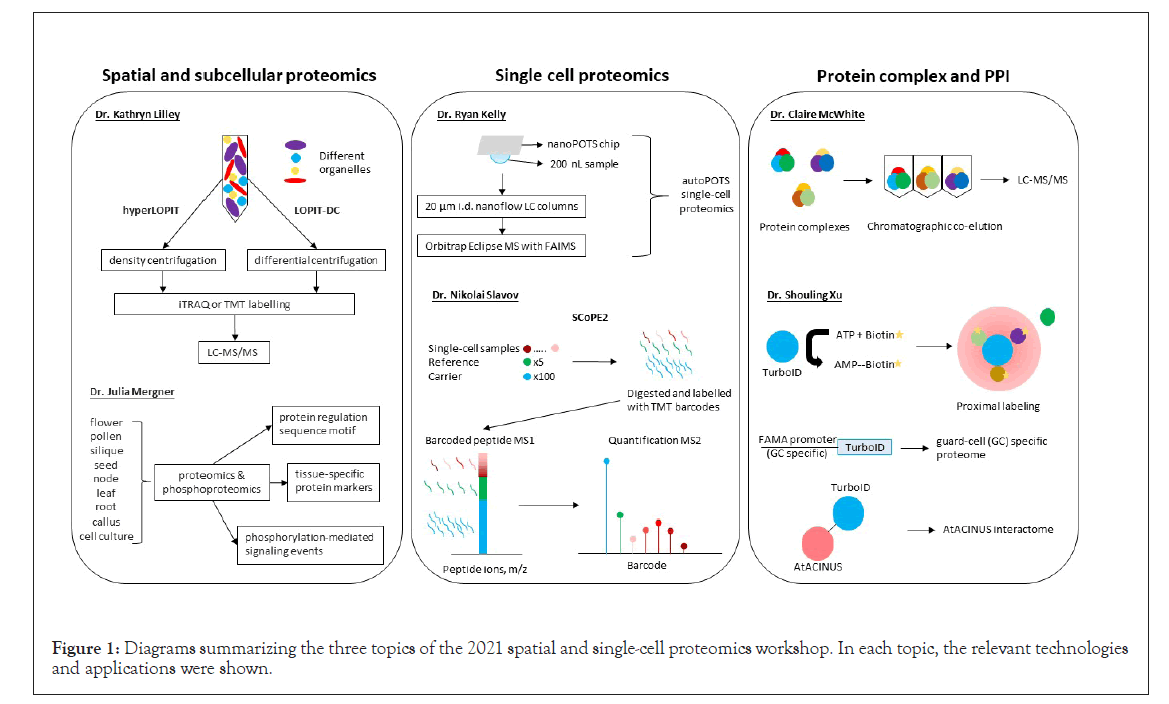
Figure 1: Diagrams summarizing the three topics of the 2021 spatial and single-cell proteomics workshop. In each topic, the relevant technologies and applications were shown.
Protein subcellular localization and compartmentation is an important aspect for eukaryotic cells. Mislocalization of proteins in cells can lead to functional perturbation and diseases. Though protein subcellular localization can be predicated mainly based on genomic and transcriptomic data, spatial and subcellular proteomics enables experimental determination of protein trafficking, dynamic and steady-state distributions in the cells, and thereby provides insights into biological functions. Dr. Kathryn Lilley’s lab from the University of Cambridge, United Kingdom, developed a protocol known as Localization of Organelle Proteins by Isotope Tagging (LOPIT). It was first used to map Arabidopsis organelle proteomes, including endoplasmic reticulum, Golgi apparatus, plasma membrane, vacuolar membrane, and mitochondria/plastids, which were fractionated by self-generating iodixanol density ultracentrifugation and then processed through isobaric tag for relative and absolute quantitation (iTRAQ)-based quantitative proteomics [1]. Protein localization was mapped via correlation of protein abundance profiles with those of known organelle or subcellular marker proteins. Dr. Lilley introduced two new technologies:
• hyperLOPIT with improved elements of LOPIT pipeline (including density centrifugation and hyperplexing with tandem mass tags (TMT), enabling high resolution of spatial proteomics [2];
• LOPIT-DC replacing density centrifugation with differential centrifugation to overcome the time-consuming and resourceintensive nature of hyperLOPIT [3].
These technologies enabled development of subcellular atlas of mouse stem cells, covering the locations of approximately 8,000 proteins and isoform-specific locations [4]. Dr. Lilley’s lab implements Bayesian mixture models for estimating the probability of a protein location and re-localization processes [5]. Now, their lab is working on phosphorylation-related localization and RNA localization in cells. Dr. Lilley has been a prominent figure in the realm of spatial proteomics and their work will continue to shape the field.
Dr. Julia Mergner’s lab from the Technical University of Munich, Germany, highlighted the lack of characterization of the Arabidopsis proteome, despite Arabidopsis serving as a model species for plant biology research. They profiled proteomes and phosphoproteomes of 30 tissue types, divided into nine groups, including flower, pollen, silique, seed, node, leaf, root, callus, and cell culture. Based on these data, they further identified tissue-specific protein markers, determined sequence motifs that function in protein regulation, and uncovered protein phosphorylation-mediated signaling events [6]. These results have been integrated into Proteomics Databank and ATHENA databases, which provide interactive access to information about Arabidopsis proteins, their modifications, and interactions.
To date, most of the proteomics studies have been on the bulk level analysis using organs and tissues, and important information in the individual differentiated cells is missed. Therefore, singlecell proteomics focusing on individual cells provides essential insights into its unique molecular components and functions, as well as heterogeneity within cell populations. Single-cell proteomics requires efficient separation of individual cells, low volume processing of single cells, and sensitive MS to analyze proteins of low abundance. Single-cell isolation techniques have been well-developed, and they include fluorescence-activated cell sorting (FACS), laser-capture microdissection (LCM), dropletbased microfluidics, and nanoliter dispensing well plates (DWP).
Dr. Ryan Kelly’s lab from the Pacific Northwest National Laboratory and Brigham Young University, USA, made seminal contributions to the development of single-cell sample processing platforms. They include nanodroplet processing in One-pot for Trace Samples (nanoPOTS) and autoPOTS, which fully automates sample preparation and analysis by integrating with the single-cell isolation techniques. Single cells isolated by FACS and LCM are dispensed on nanoPOTS chips, droplet-based nanoliter DWP, where proteins are digested in nanodroplets, and then taken into liquid chromatography (LC)-MS/MS [7]. The nanoPOTS chip requires only 200 nL samples with 0.8 mm2 surface area, which greatly improves enzyme kinetic efficiency [8]. Both nanoPOTS and autoPOTs offer capabilities for low-input single cell proteomics. Their recent work is to improve protein identification by applying 20 μm i.d. nanoflow LC columns coupled with Orbitrap Eclipse MS with field asymmetric ion mobility spectrometry (FAIMS) technique. This optimization has improved the identification of protein groups from 211 to around 1200 in single cell experiments. The inclusion of FAIMS enhances proteome coverage in single cells because it filters out the singly charged ions before they enter the MS to increase sensitivity. Finally, the improvement of data analyses from MaxQuant to Proteome Discover further increased the number of identified proteins.
Dr. Nikolai Slavov’s lab from Northeastern University, USA, reported the development of another single-cell proteomics approach, i.e., Single Cell ProtEomics by Mass Spectrometry Version 2 (SCoPE2). In SCoPE2, single-cell samples, reference (5 cells) and carrier (100 cells) are labelled with iTRAQ or TMT reagents. This approach takes advantage of multiplexing with carrier cells to enhance MS1 signal of precursor peptides, and peptide quantification from single cell samples can be resolved at MS/MS level. SCoPE2 increases protein quantification throughput at a low cost. Integrative analysis with transcript data showed that protein copies per gene quantified by SCoPE2 were 20-fold more than RNA copies by 10 × Genomics. Thus, the high copy number of proteins from single cells lays the foundation for further inferring transcriptional and post-transcriptional regulations [9]. It should be noted that the quantitative analysis may be compromised by the carrier group precursor contamination/co-isolation. Although the SCoPE2 approach is different from that presented by Dr. Kelly, both advancements have laid the foundation for large-scale single cell proteomic studies in the near future.
Protein complex formation and PPIs are important processes in cellular signaling and metabolic pathways. Identification of the interacting proteins and proximal neighbors are key to understand protein functional context and inform its subcellular localization. In recent years, a suite of MS-based methods has emerged capture potential protein complexes and PPIs, including affinity purification MS (AP-MS) and proximity labeling (e.g., biotin-labeling based BioID and TurboID, as well as pupylationbased interaction tagging (PUP-IT)). Dr. Claire McWhite’s lab from Princeton University, USA, developed a protocol known as co-fractionation MS (CF-MS), which does not require any antibodies or epitope tagging. This protocol is based on the principle that proteins with physical association in a complex will co-elute in chromatography [10]. Native protein extracts can be chromatographically fractionated by traditional methods such as size exclusion chromatography, ion exchange chromatography, isoelectric focusing, glycerol gradient separation, etc. The separated protein groups are digested with trypsin, identified, and quantified by LC-MS/MS. By using CF-MS, they successfully verified 117 protein complexes from 13 plant species (McWhite et al.). In this study, they also implemented machine-learning to identify sets of co-occurring proteins (using a model trained on known interactions) and later validated the output by matching phenotypes as malfunction of a protein complex may lead to changes in phenotype. Additionally, they were able to predict novel phenotypes based on protein interactions.
Dr. Shouling Xu’s lab from Carnegie Institution for Science, USA, shared their research on mapping PPI through proximitylabeling MS (PL-MS). Their lab focuses on using biotin ligase to covalently bind biotin to neighboring proteins, which can then be purified from total protein extracts using streptavidin beads. The biotinylated proteins are then identified using LCMS/ MS. This technology is called BioID. TurboID is a modified biotin ligase, which has higher activity than BioID and thus requires less biotin-incubation time [11]. Dr. Xu showed some examples of PL-MS applications. For example, they mapped guard-cell specific nuclear interactome by fusing guard-cell specific promoter of FAMA with TurboID and split Venus [12]. FAMA is a basic helix-loop-helix transcription factor involved in guard cell development. In addition, they verified Arabidopsis AtACINUS (AT4G39680) was an O-glycosylated regulator of transcription and alternative splicing during reproductive transitions by mapping its interactome using TurboID [13]. Their future works include construction of inducible split-TurboID and mapping organelle-specific proteome by fusing organelle-specific marker protein with TurboID. These new molecular tools have substantially expanded the utility of the PL-MS and can be customized to investigate protein-protein interactions in specific cell-types and ultimately in a single cell [11-13].
The last session of the workshop was a panel discussion. The discussion was focused on current technologies, their accessibility and future development. Currently, the research community is working diligently to further technological advancement and increase its accessibility for the community. For example, efforts have been made to simplify single-cell sample processing in order to enable regular laboratories to conduct single-cell proteomics without the need of sophisticated nanoPOTS. Another point of discussion was the future of proteomics and possible merging of different technologies, such as identifying protein complexes in single-cell proteomic analysis. Protein enrichment strategy remains a hot topic in proteomics research at the single-cell level. Besides technical advances and opportunities, the panel speakers discussed bioinformatic data processing, statistical challenges, and false discovery rate in high resolution proteomics. Three emerging directions in current proteomics have emerged from the discussion:
• Improving technology and methodology for increasing sensitivity and coverage of proteomic analysis;
• Updating existing and developing new computational tools in accordance with the technology requirement; and
• Increasing accessibility of new tools to a wide range of laboratories.
Future perspectives
Proteomics is a fast-advancing field. Its applications to many areas of biological and medical research have provided important insights into different biological processes that are not fully understood. This workshop uncovered cutting-edge technologies and research frontiers in proteomics, specifically in spatial and subcellular proteomics, single-cell proteomics, and proteinprotein interactions. Each speaker discussed the spatial proteomic approaches that either focused on subcellular and single-cell resolution or enhanced proteome coverage. Additionally, a shared goal was to make the technologies accessible and affordable for the research community. The proteomics community expects to continue improving sample preparation to minimize sample loss and enhancing MS sensitivity for single cell, as well as detection and identification of protein modifications and/or protein complexes in very low quantity or stoichiometry that are present in single cells or subcellular compartments. Furthermore, there are striving efforts to map the protein atlas and PPI networks of organelles in the cell using PL-MS, such as TurboID, where specific markers of each organelle may be used. As new proteomics technologies (e.g., LOPIT-DC, SCoPE2 and PL-MS) are further improved and become more readily available, great discoveries and advancements in basic and applied research can be expected.
Citation: Lin C, Pelosi J, Vela S, Chen S (2021) Spatial and Single-Cell Proteomics Workshop Report. J Proteomics Bioinform.14:553.
Received: 21-Sep-2021 Accepted: 05-Oct-2021 Published: 12-Oct-2021 , DOI: 10.35248/0974-276X.21.14.553
Copyright: © 2021 Lin C, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.