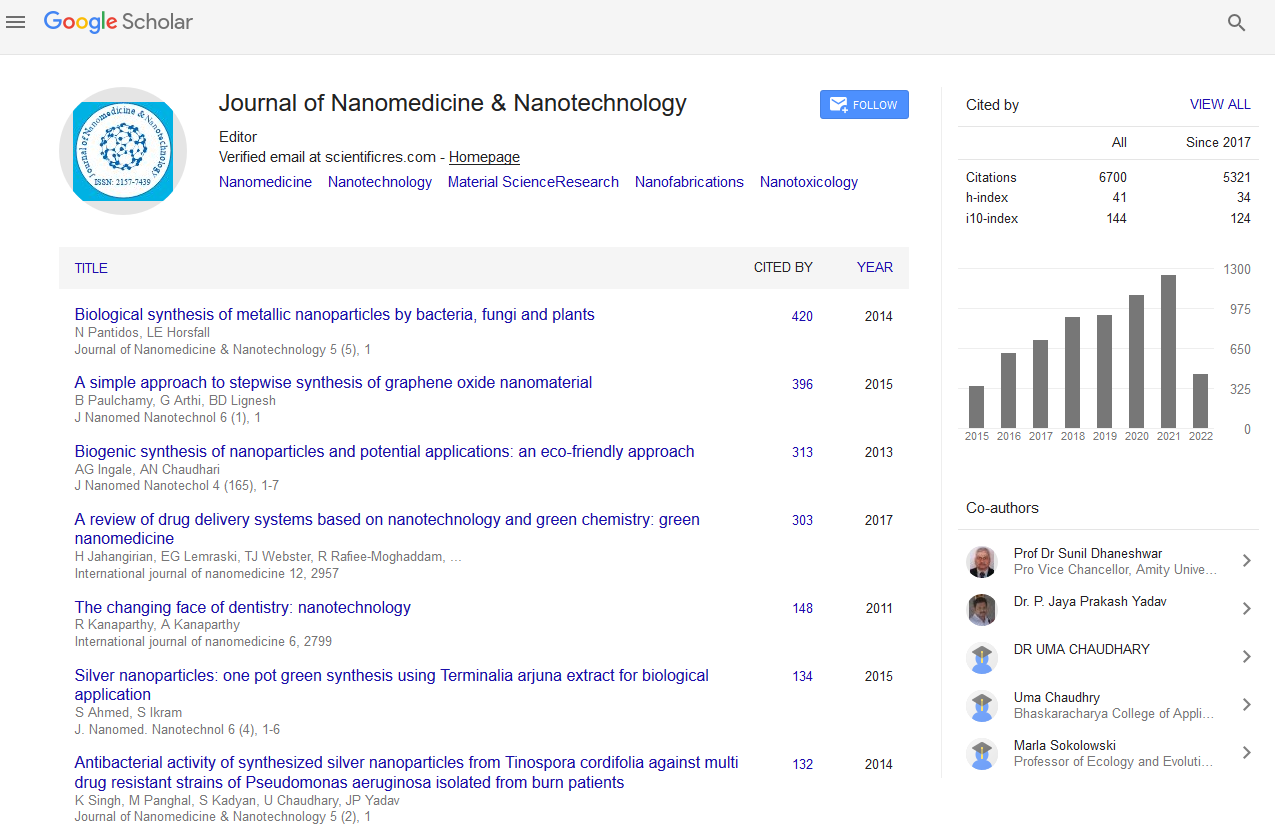
PMC/PubMed Indexed Articles
Indexed In
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- China National Knowledge Infrastructure (CNKI)
- Electronic Journals Library
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- MIAR
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Catabolic genes dependent detection of aerobic BTEX degraders
7th World Congress on Petrochemistry and Chemical Engineering
November 13-14, 2017 Atlanta, USA
B Icgen and A Yavas
Middle East Technical University, Turkey
Posters & Accepted Abstracts: J Pet Environ Biotechnol
Abstract:
Monoaromatic hydrocarbons, including benzene, toluene, ethylbenzene and xylene, collectively called BTEX, are major components of gasoline and are thought to be the most significant contaminants of soil and groundwater due to frequent leakages from underground storage tanks and accidental spills. Degradation of BTEX compounds by bacteria is known to be one of the most efficient ways to remove these compounds from soil and groundwater. There have been extensive studies demonstrating that BTEX degradation is performed by variety of pathways including the oxidation of methyl group, ring monooxygenation at positions 2, 3, or 4 or ring dioxygenation using the toluene as a model hydrocarbon. In lower pathway, ring cleavage is mediated by catechol dioxygenases after which the molecule is further degraded into citric acid cycle. Therefore, this study aimed at screening of aerobic BTEX degraders with their corresponding catabolic genes xylA (291 bp), todC1 (510 bp), tmoA (505 bp) and catA (282 bp) by ploymerase chain reaction (PCR). Methodology & Theoretical Orientation: 20 different bacteria previously isolated and identified by our lab from river water contaminated with petroleum hydrocarbons, including the strains of Pseudomonas plecoglossicida Ag10, Raoultella planticola Ag11, Staphylococcus aureus Al11, Staphylococcus aureus Ba01, Stenotrophomons rhizophila Ba11, Delftia acidovora Cd11, Staphylococcus warneri Co11, Pseudomonas koreensis Cu12, Acinetobacter calcoaceticus Fe10, Pseudomonas koreensis Hg10, Pseudomonas koreensis Hg11, Staphylococcus aureus Li12, Serratia nematodiphila Mn11 Acinetobacter haemolyticus Mn12, Comamonas testosteroni Ni11, Acinetobacter johnsonii Sb01, Pantoea agglomerans Sn11, Micrococcus luteus Sr11, Micrococcus luteus Sr11, Acinetobacter haemolyticus Zn01, were used in this study. Bacteria were routinely grown in nutrient broth for DNA extraction. Total DNA extraction was done by alkaline lysis method with some modifications. Isolated DNA was used as a template and PCR was carried out by using the xylA (side chain monooxgenase), todC1 (ring hydroxylating dioxygenase), tmoA (ring hydroxylating monooxygenase) and catA (ring cleavage dioxygenase) primer sets in a 25 µl reaction mixture containing 50 ng template DNA, 5 pmol forward and reverse primers, 100 µM of each dNTP and 2.5 µl NEB 10X Taq reaction buffer and 1.25 U Taq DNA polymerase (NEBM0230). Finally, PCR products were run on 1% agarose gel and visualized under UV light by using quick-load 100 bp DNA ladder (100-1517 bp) as marker. Conclusion & Significance: Upper pathway genes xylA, todC1, tmoA and lower pathway gene catA were tried to amplify by PCR. Ring-hydroxylating dioxygenase and catechol dioxygenase genes were detected in 15 and 16 different bacteria respectively. No ring hydroxylating monooxgenase gene was amplified by PCR. The xylA gene encoded on TOL plasmid and responsible for the sidechain monooxygenation was not detected among species. These results indicated that bacterial isolates used in this study likely use dioxygenation pathway (todC1) in the initial attack of aerobic BTEX degradation and utilize catechol 1, 2 dioxygenase (catA) for ortho cleavage of the aromatic ring.
Biography :
B Icgen is a Faculty Member at METU in the Department of Environmental Engineering. His interests lie in the fascinating and often complex array of processes taking place in microbial environment and the behavior of the microorganisms under different environmental conditions. His team focuses on microbial biotechnology, with a recent focus on environmental genomics, transcriptomics and proteomics.