PMC/PubMed Indexed Articles
Indexed In
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Geneva Foundation for Medical Education and Research
- Google Scholar
Useful Links
Share This Page
Journal Flyer
Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Review - (2019) Volume 8, Issue 3
Tripartite Motif Cofactors, a Novel Gene Target for Liver Cancer via Regulating the Immune Cells and Gut Microbiome: A Review
Ifeoma P Okoli1,2*2Department of Pharmacology and Therapeutics, Umuna Orlu Campus, Imo State University, Owerri Imo State, Nigeria
Received: 14-Aug-2019 Published: 14-Oct-2019
Abstract
TRIM, a multi-domain protein associated with N-terminal ring finger E3 ligase and C-terminal plant homeodomain/ bromodomain PHD chromatin interacting module, N-terminal ring finger known as ring B-boxes and coiled-coil RBCC domain with structure that underscores biochemical reaction which requires enzymes like E1, E2 and E3 of which E3 serves as receptor recognition for target proteins. Most TRIM proteins are E3 ligases in the ubiquitination cascade that translated in diverse physiological and biological processes such as differentiation, growth, transcription and oncogenesis. Implicated in pathological processes from Mendelian inherited disorders, cellular (plethora) processes like cell cycle regulation, innate immune response and apoptosis to cancer. Genetic factors are at high risk and contributed between 30%-50% disease prevalence like obesity, cirrhosis. TRIM28 (TIF1β), TRIM24 (TIF1α), TRIM33 (TIF1γ) are cofactors of tripartite motif TRIM subfamily protein, distinct transcriptional factors that correlate with each other and interact with other proteins both in functional and physical in cancer disease. Studies have shown that TRIM protein is a regulator in inflammatory, infectious and cancer diseases. This review focused on tripartite genes as a liver cancer target via regulating immune cells and the gut microbiome. More on research so far, disease development, progression and influence. Considering the incident rate and progression, genetic involvement remain challenging thus needs more insight on prognosis that will potentiate clinical effect with lesser adverse events and recurrences that will benefit the patients.
Keywords
TIF1αβγ; Bile acids; Lipopolysaccharides; Pattern recognition receptors; Immune cell; Inflammation
Abbreviations
TRIM: Tripartite Motif; ALD-Alcoholic Liver; NAFL: Nonalcoholic Fatty Liver; NASH: Nonalcoholic Steatohepatitis; PHD: Plant Homeodomain/Bromodomain; RBCC: Ring B-Box and Coiled Coil; KRAB-Znfs: Kruppel-Associated Box Domain Zinc Fingers; KAP1-KRAB: Associated Protein 1; PBC: Primary Biliary Cirrhosis; AIH: Autoimmune Hepatitis; GWAs: Genome-Wide Association Studies; H3K9me3: Histone-3- Lysine 9 Trimethylation; IPSCs: Induced Pluripotent Stem Cell; KLF4-Kruppel-Like Factor 4; NR-Nuclear Receptor; AF-2: Activated Function-2; MSK1: Mitogen-And Stress-Activated Protein Kinase; FSP27: Fat Specific Protein 27; HCMV: Human Cytomegalovirus; ATM: Ataxia Telangiectasia Mutated; PAF: Pol 11-Associated Factor; POL 11: Pausing of RNA Polymerase 11; ULU: Upper Limit of Normal; ALT: Alanine Transaminase; ALP: Alkaline Phosphatase; BBV: Blood-Borne Virus; SNP: Single Nucleotide Polymorphism; aCGL: Array Comparative Genomic Hybridization; CNV: Copy Number Variation; TFs: Transcription Factors; RARA: RAR α Receptor; NF-kB: Nuclear Factor Kappa Beta; PRR-Pattern Recognition Receptor; TLR-Toll-Like Receptor; MDR2-Multi Drug Resistance Protein 2; CBA-Conjugated Bile Acid; SIPR2-Sphinogosine 1-Phosphate Receptor 2; TcF/LEF-T-Cell Factor/ Lymphoid Enhancer Factor; GSK2β: Glucose Synthase Kinase 3 Beta; APC: Adenomatous Polyposis Coli; TKR: Tyrosine Kinase Receptor; TGFβ: Transforming Growth Factor; RB1: Retinoblastoma B1; IGF2R: Insulin-Like Growth Factor 2 Receptor; hESCs-Human Embryonic Stem Cell; PNPLA3: Patatin-Like Phospholipase Domain Containing 3 Protein; HSD17B13: Hydroxysterol 17-Beta Dehydrogenase 13; CYP7A1: Choleterol 7α Hydroxylase; CYP7B1: Cholesterol 7α Hydroxylase β1; CYP27A1: Sterol 27 Hydroxylase A1; OCA: Obeticholic Acid; CDCA: Chenodeoxycholic Acid; MCA: Muricholic Acid; FXR: Farnesoid X Receptor; CA: Cholic Acid; TCA: Taurocholic Acid; GCA: Glycocholic Acid; TCDCA: Taurochenodeoxycholic Acid; GCDCA: Glycochenodeoxycholic Acid; UDCA: Ursodeoxycholic Acid; DCA: Deoxycholic Acid; LCA: Lithocholic Acid; FGF: Fibroblast Growth Factor; BDA: Bile Duct Obstruction; ERK1/2: Extracellular Signal-Related Protein Kinase 1/2; T2D: Type 2 Diabetes; GPCRs: G Protein Coupled Receptors; Atg16L1: Autophagy Related 16 Like 1; SARM: Senile Α- and Armadillo-Motif; IRF3: Interferon-Regulatory Factor 3; TIR: Interleukin 1 Receptor- Domain-Containing Adaptor Protein; TRIF-TIR Domain-Containing Adaptor-Inducing Interferon B; PAMP: Pathogen Associated Molecular Pattern; MAMP: Microbe Associated Molecular Pattern; LPS: Lipopolysaccharide; LTA: Lipoteichoic Acid; IL: Interleukin; IFN: γ-Interferon Gamma; SIBO: Small Intestinal Bacterial Overgrowth; CTL: Cytotoxic T Lymphocyte; CGZB: Cytotoxic Granzyme B; BCL-2: B Cell Lymphoma 2; TNF: Tumor Necrosis Factor; CMML: Chronic Myelomonocytic Leukemia; Cvt: Cytoplasm-to-Vacuole Targeting; α-TEA: Alpha-Tocopheryloxy Acetic Acid; NPC: Non-Parenchymal Cell; HSC: Hematopoietic Stem Cell; TSP: Tissue Specific Phenotype; C/EBPβ: CCAAT Enhancer Binding Protein Beta; STAT3: Signal Transducer and Activator of Transcription 3; AP-1: Activator Protein 1; LrNK: Liver Resident Natural Killer Cell; Treg: Regulatory T Cell; FOXP3: Forkhead Box P3; STARTRAC: Single T Cell Analysis By RNA-Seq and TCR Tracking; TME: Tumor Microenvironment; ATM: Anti Tumor Immune; HMGB1: Membrane Bound High Mobility Group B1; HLA-C1: Human Leukocyte Antigen–C Group 1; CNV: Copy Number Variant; KOX1/ZNF 10: Human Zinc Finger Factor 10
Background
Liver being important organ in the body, perform great function yet stand-alone, in disease state fear troupe in and one cannot mention liver cancer without the diseases of the liver. Liver cancer (cancer that starts in the liver) are disorders that begin in the cells of the liver, those that begin in other areas of the body and spread to the liver are termed metastatic. Hepatitis ABC, cirrhosis and alcoholic are leading in liver cancer, which ranked three high causes of death [1-3]. Liver cancer disorder, face with two complexes histologic pattern [4]. Clinical features like alcohol and nonalcoholic fatty liver that includes NAFL (fatty liver without damage) and nonalcoholic steatohepatitis NASH (obesity, lipid metabolism disorders, insulin resistance). That presented with histological activity in both grade and stage (fatty liver and steatohepatitis) and clinical association specifically on etiology (insulin resistance, lipid, alcohol and drugs) with natural history and mechanism varies [4].
Classification of liver diseases seems difficult because no best way to classify fatty liver disorders, with guidelines on recent research articles can distinguish based on overlapped syndrome between PBC and AIH [5]. This report suggested that implication for AIH therapy, not at classification point alone. However, fatty disorders are been classified as alcohol and nonalcoholic but not known if the pathologic condition of overlapping expression represents steatohepatitis and other criteria [4,6].
Globally, liver diseases have recorded the highest death rate and there is a need to identify the key gene that plays roles in its onset and development. Common types are hepatocellular carcinomas develop from liver cells (hepatocytes) which present with cirrhosis, viral infections [1,2,7], and steatohepatitis. Cirrhosis, hepatitis B, C virus (HBV, HCV) which tends to be most deadly among all other types and cholangiocarcinoma develop from the bile duct, although the rate of progression varies from individual. Gene disorders and metabolic syndrome remain the key genesis of this deadly disease [8-11]. Genetics factors are at high risks, which contributed 10-30% heritability in GWAs and account to between 30-50% in disease prevalence like obesity, cirrhosis, T2DM [12].
Genome-wide association study identified large amount of genetic variant associated with various human disease that cluster in families, these were present with heritability risk loci provided on a sample number in a population [13] To explain genetic influence on common diseases and heritability in human disease-associated variant mostly occur in protein coding region than on genotyping array. Of which various compatible approaches has developed to incorporate analysis of CNV in design array and linkage disequilibrium relationship between common copy number polymorphism CNPs and SNPs data [13-16].
Tripartite motif family protein classifies into eleven subgroups of C-1-X1, among all TRIM24, TRIM28, TRIM33 are in the same subgroup C-VI [17-19]. Emphatically, a subgroup of tripartite motif TIF1β is in-relation with TIF1α, though TIF1γ on which the genetic influence identified [20,21]. Notably, interferon IFN upregulate the expression of the various TRIM gene and trigger an antiviral response. However, are not induced by IFN in some and were not detected in few others like TRIM1, TRIM11, TRIM28 and TRIM62 [18]. Remarkably, in human monocyte-derived macrophages, interferon type 1 downregulated the expression of TRIM28, whereas type 2 induces downregulate of TRIM28 [18]. Other study observed cross-link of Fϲγ by immune complex IC which negatively regulate IFN-induced signaling, in the sense IC-mediated mechanism inhibited by IFN gamma signaling [22], whereas TRIM9 and TRIM54 upregulated in fc region of IgG receptors FϲγR-activated macrophage [18]. Furthermore, the study identified risk loci as PNPLA3 for ALD and TM6SF2, also MBOAT7 [23,24], considering their combined effect as a genetic risk factor for NAFLD [25]. Moreover, TRIM 28 and a subset of KRAB-ZNFs identified a regulator of DNA methylation on reprogramming as KRAB/ZNFs interact with TRIM28 to induce H3K9me3 DNA methylation [26-28]. Identified as a repressor, modulator [21,29], coactivator [30], DNA repair [20], tumor suppressor [20] and corepressor [31-34]. Noteworthy, kruppel-like factor 4, a protein known for pluripotency and reprogramming induced pluripotent stem cell, differentiation depends on cell status which represses gene expression, thus involve in various pathway together with TRIM28 by modulating H3K9me3 DNA methylation and somatic cell are reversible [26].
TRIM24 interact with the ligand-binding domain, a different NR ‘retinoic acid’ receptors RAR [35], mediator of AF-2 to chromatin remodeling [36] and Msug1 [37], of which RA signaling altered in HCC developed TRIM24 knockout mice [38]. Remarkably, major pathway for liver carcinogenesis implicated by p53 inactivation via TP53 mutation gene, activation of Wnt-β catenin pathway via CTNNB1/β-catenin, have both positive and negative regulatory role on AXIN1 [39-41]. Thus knockout mice genetics showed an increase of HCC which are significantly implicated (RARA) in liver-specific cancer [38]. Importantly, the study has shown a familial clustering at an early age of onset of liver cancer developed to HCC and major gene involved [42,43]. Mendelian autosomal recessive major genes identified to a play role in HCC etiology [42]. Thus, TRIM24 inactivation promotes tumorigenesis showing cellautonomous process [20,21].
TRIM28 interact with kruppel-related zinc-finger transcription factor known for DNA repair and transcription regulation. TRIM28 acts as a tumor suppressor whereas function as oncogene [20,21]. DNA damage recognition by ATM and initiation of genotoxic stress response [44,45]. Also in response to recognition and repair of DNA-damage that modulated by heterochromatin protein 1 to histone, suggests its involvement in DNA damage response pathways [34,46,47]. Consistent with ATM- and KAP1- independent pathways of chromatin relaxation in the DNA damage response may have been from phosphorylation of serine 824 [44]. The KAP1 localization at sites of DNA strand break response to ionization radiations, signaling pathway could be the perfect way of ATM- and KAP1- in DNA repair [34]. Recently study identifies TRIM28 (KAP1) role as cofactor and regulator to myoblast differentiation MyoD function of which phosphorylation of KAP1 mediated by MSK1 thereby releases corepressor from the scaffold [48]. A heterochromatin-induce activity of KAP1 was suppressed by mTOR-mediated phosphorylation driven by KAP1 knockdown of latency either by activation of NF-KB with immune-based drug and ATM combination [33]. TRIM28 have increased tendency to tolerate environmental stress thus serves as link between gut microbiota and liver cancer which FXR inactivation, increase BAs levels contributed to inflammation and tumorigenesis in TRIM28 hepatocytes [49]. Environmental factors as diet and infection are main element involved diseases of liver, noting the closure of the microbes live in the intestine. Gut microbiota play roles in chronic inflammation of liver disease thus influence liver phenotype in various ways. Gut microbiota activated TLR/NLR that motivate expression of pro-inflammatory gene thereby promote liver inflammation [50]. Knockdown of KRAB-associated protein 1 KAP1 (liver-specific) showed similarities at FSP27 gene site and detoxification, of note male are particularly being challenged [31]. It is essential KAP1 identified as corepressor that phosphorylate on serine 824 human cytomegalovirus infect cells thus inhibited by mammalian target rapamycin mTOR, effect enhanced by addition of NF- KB inducer (TNF) [33]. HCMV latency may have driven by pharmacological activation of ataxia telangiectasia mutated (KAP1).
TRIM33 interact with SMAD4 act as tumor suppressor gene of chronic myelomonocytic development plays a role in preventing apoptosis in B cells lymphoblastic leukemia [51]. The study has been reported that trim 33 act as an oncogene [51-53], although depends on the cell type and as tumor suppressor [54-56]. Various researches have reported its involvement in blood cancers [51,55,57], which was shown to interact with TRIM24. Study observed loss of TRIM- /- prevent apoptosis thereby blocked the enhancer-mediated Bimactivation and functions as a negative regulator to the activity of the enhancer [51]. In addition, loss of function of pausing of RNA polymerase II-associated factors PAF in trim gamma-deficient mice, which reduced hematopoietic stem cells HSC function and blood defects [58,59]. Deficient embryos of trim 33 was unable to give responses to inflammatory recruitment signals at the site of infection and decreased basal motility showing involvement in inflammation in both macrophages and neutrophils [60]. Accordingly, PHD-bromodomain identified, which binds histone H3 tail in order to recruit trim 33 to the chromatin and E3 ligase stimulation, also have ability to ubiquitinate Smad4 complex at TGF-β superfamily [61]. Furthermore, loss of trim33 was identified in chronic myelomonocytic leukemia CMML mice, although could not detects mutation in the coding sequence of gene which may be as a result of the reduced expression in 35% patients with CMML that goes almost undetected [55]. The expression of Trim33 restores monocytes in a patient who respond to demethylating agent decitabine. Furthermore, trim 33 prevented apoptosis in B lymphoblastic leukemia, achieve by blocking activation of enhancermediated Bim (proapoptotic protein gene), also recruited by Pu.1 yet antagonize its function [51]. Importantly, significant deletion of the region at chromosome 1p13.1 in the myelomonocytic patient was also reported [62]. Despite the small size of aberration, deletion 1p has reported for showing most abnormalities of which have also identified with trim 33 at 1p13.1 thus seen in myelomonocytic patients [56,63,64]. Trim 24 and 33 showed to form a complex that in turn suppress tumor in HCC (Figure 1) [20,21].
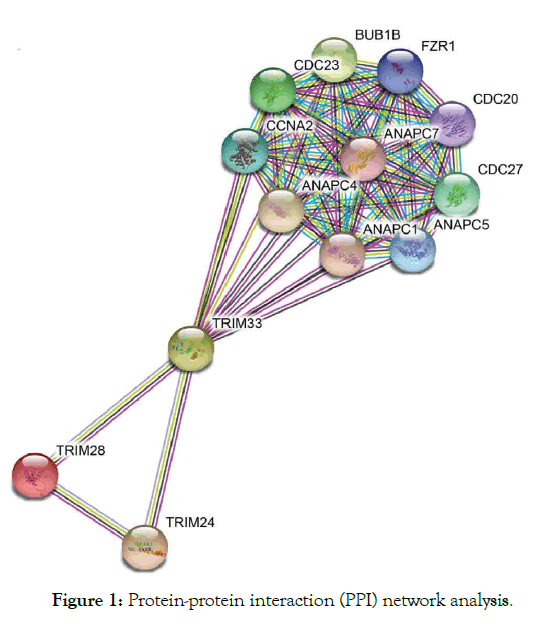
Figure 1: Protein-protein interaction (PPI) network analysis.
TRIM sub-family proteins: C -terminal domain
TRIM, a multi domain protein associated with N-terminal (ring finger E3 ligase) and C-terminal (bromodomain PHD chromatin) interacting module known as B-boxes and coiled coil domain with structure that underscores biochemical reaction. TRIM 24,28,33 are three member family that have PHD signature and nuclear protein which forms multimeric complexes for transcription and remodeling and distinct transcriptional factors that correct with each other and interact with other proteins both functional and physical in cancer diseases [20].
Tripartite motif family proteins classify into 11 subgroups of C-1 - X1, among all Trim24, Trim28 and Trim33 are in the same subgroup C-V1 (Figure 2).
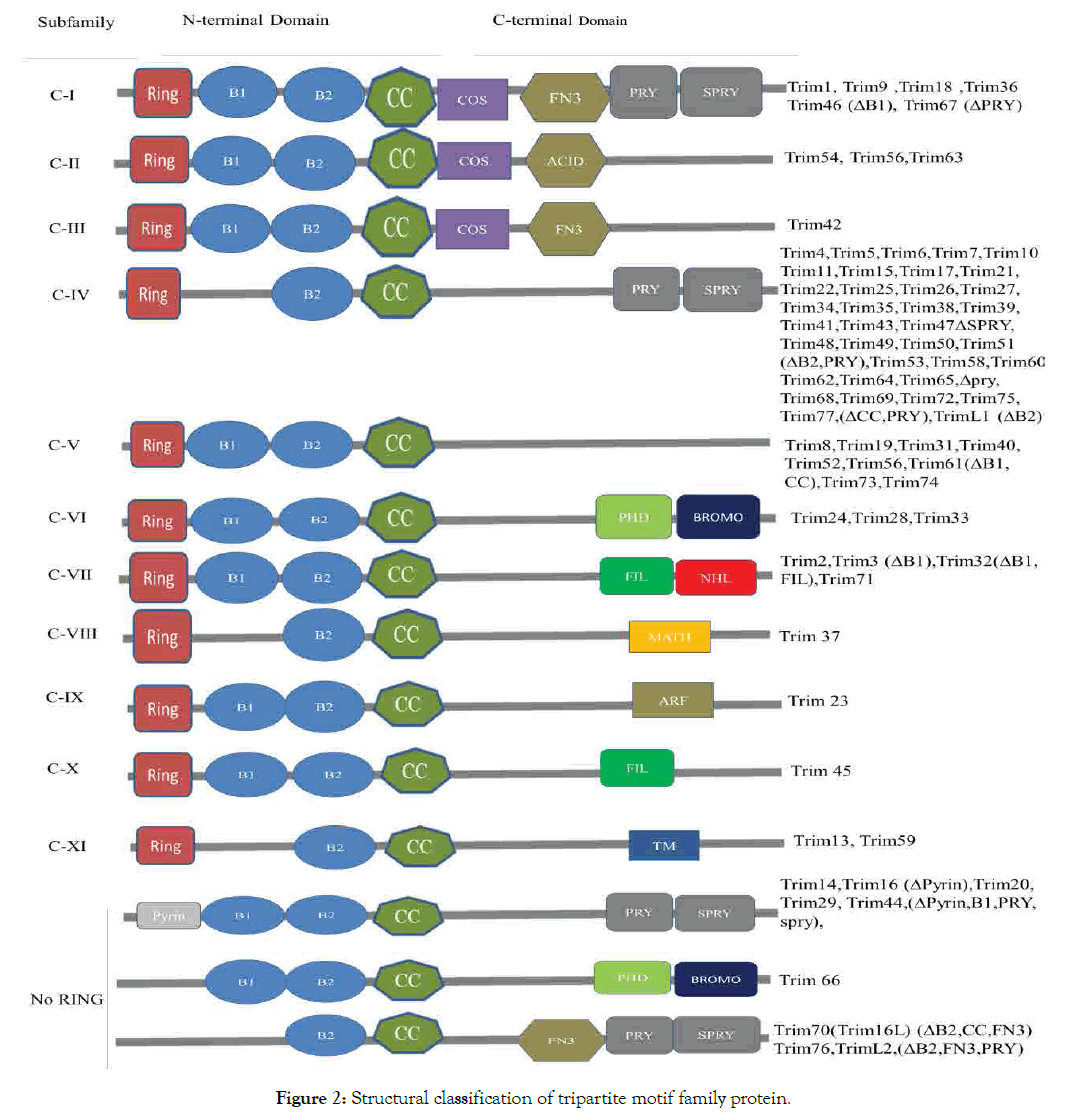
Figure 2: Structural classification of tripartite motif family protein.
The risk factors and current mechanism of the liver cancer
The risk factors:
Cirrhosis: Cholangitis as formerly known refer now PBC, an autoimmune disease of liver [65]. Cholestasis liver present symptoms like fatigue [66], pruritus [67], portal hypertension [68], hyperlipidemia [69], bone disease [70], vitamins deficiency [71]. Classical features causes intrahepatic bile duct damage, inflammation of portal and periportal (interface hepatitis) and later to fibrosis (fibrous septa) with three scores (portal, bridging and perisinusoidal fibrosis) of complex features eventually develop (as scarring) to cirrhosis [4,65,72,73]. Further study report its development from cholestasis to fibrosis and finally to cirrhosis [74]. The PBC prevalence showed an increase in families with affected member possibly environmental factors such as chemical and infection can induce PBC [75,76]. Cholangiocytes is the cells involved in cholestasis liver diseases representing clinical features as PBC that may progress to autoimmune hepatitis not significant; some patient may present with both PBC and AIH. Other features base on cholestasis serum liver test, outline of positive non-cholestasis liver injury. PBC usually presented with antinuclear antibodies ANA, pathology of intrahepatic bile duct in 90-95% patients [73,77], anti-smooth muscle antibody ASMA test (increase immunoglobulin IgM and hyperglobulinemia IgG) and histology presenting features such as elevated upper limit of normal in ALT, ALP, anti-mitochondrial antibody AMA (negative/positive) [73]. Given that AMA presented in 95% of PBC patients [78], of which magnitude of biopsy (showing bile duct and non-suppurative cholangitis damage) sample is necessary for diagnosis. Treatment with immunosuppression and ursodeoxycholic acid UDCA are normally used to aid autoimmune overlap syndrome in diagnosis [5,79]. Given that risk of reduction of pruritus in UDCA therapy with rifampin showed no significant in PBC patients other than rifampin associated with hepatotoxicity [67].
Treatment of PBC based on elevated alkaline phosphatase level is the current diagnosis, as ursodiol was only treatment approved for primary biliary cirrhosis, thus concomitant administration with obeticholic acid OCA showed decrease alkaline phosphatase and bilirubin level baseline [80]. Other study indicated a significant increase in reactive oxygen specie ROS and apoptosis in hepatocytes in Ghr-/- and Mdr2-/- knockout mice compare to control, thus showed highly downregulation in hepato-protective gene of Hnf6, Egfr [81]. Expectedly, an increase in serum material associated with liver damage and cholestasis in Ghr-/- and Mdr2-/- knockout mice, increase collagen deposition and bile duct proliferation related to Mdr2-/- knockout mice [81]. Importantly, link between HLA allele and PBC has been reported [82-84], risk allele occur in gene associated with immune function that intersect in various immune pathways, which identified in GWAs study as B, T, myeloid cell differentiation and antigen presentation [85]. Noting that three loci gene in TNF-α signaling pathway identified by GWA study: TNFRSF1A, DENND1B, TNFAIP2 [82,86]. Study revealed of insulin resistance and cirrhosis in NAFLD patients compared to control with a familiar clustering and potential maternal linkage [12,87]. Another study reported of HCV related with odd ratio 17; 95% Cl, 4.2-12.9 significant increase prevalence of cirrhosis among first degree patients relative [88], also other genetic evidence of linkage in patients with HBV related HCC [89]. In addition, identified PNPLA3 [90], TM6SF2 [91], SAMM50 [92], ERLIN1 [93] association in liver disease. Recently studies also identify SLG39A12 encoding solute carrier family 39 member12, GPT and GOT1 encoding AST and ALT respectively [94].
Profoundly, PBC recurrent reported in 25% of patients, which occurred after liver transplant [73]. Liver biopsy is needed for diagnosis of post-transplant to certain AMA persistence, thus preventive treatment reduces risk of recurrent with UDCA after transplant although not confirm yet [95]. Whereas treatment other than clinical and characteristics risk factors has been confirmed. Study have established that chronic liver inflammation motivated by HCV, HBV, NAFLD and alcohol intake awaken by HCC should targeted at early detection, prevention and surveillance [96], which can restrict and activate immunity of neoantigen-specific T cells in immunotherapy [97], and inflammation associated as key hallmark of cancer (Figure 3) [98].
Figure 3: Stages of primary biliary cirrhosis.
Hepatitis B and C: Hepatitis is a disease that affects the liver, which threatens the liver ability to function having different symptoms and treatment. Various types are A, B, C, D and E, laboratory tests can determine hepatitis types. Causes include virus, recreational drugs, injury and contact with infected person. Here we are going to focus on hepatitis B and C types that tend to be more dangerous than others are. Hepatitis B spread mainly through blood contact with infected or mother to infant delivery, which can lead to serious health issues like cirrhosis that risks related to age. Acute hepatitis B virus HBV estimated at 20,900 cases, while chronic HBV estimated at 2.2 million cases in United States 2016 [99,100]. Hepatitis C is a blood-borne virus spread by sharing sharp objects and drugs injecting equipment. Chronic HCV also can be acute or chronic even death. Acute HCV estimated at 41,200 cases, which can become chronic in 75-85% cases [100,101], people living with HCV estimated at 2.4 million in 2013-2016 in United State [102,103], histological stage and advanced fibrosis score in PBC associated with HBV infection [104]. Studies suggested that some host factors such as immunity could be acquire to protect against viral HCV persistence [105], and reinfection risk after repeated clearance associated with previous infection and HCV-specific T-cells [106]. Suggest that vaccine for spontaneous clearance more preferable than vaccine for primary HCV prevention (Figures 4 and 5).
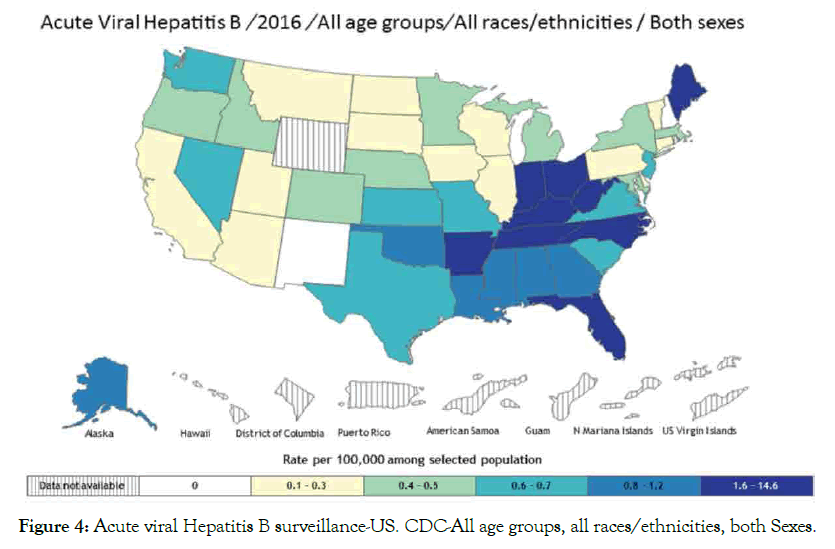
Figure 4: Acute viral Hepatitis B surveillance-US. CDC-All age groups, all races/ethnicities, both Sexes.
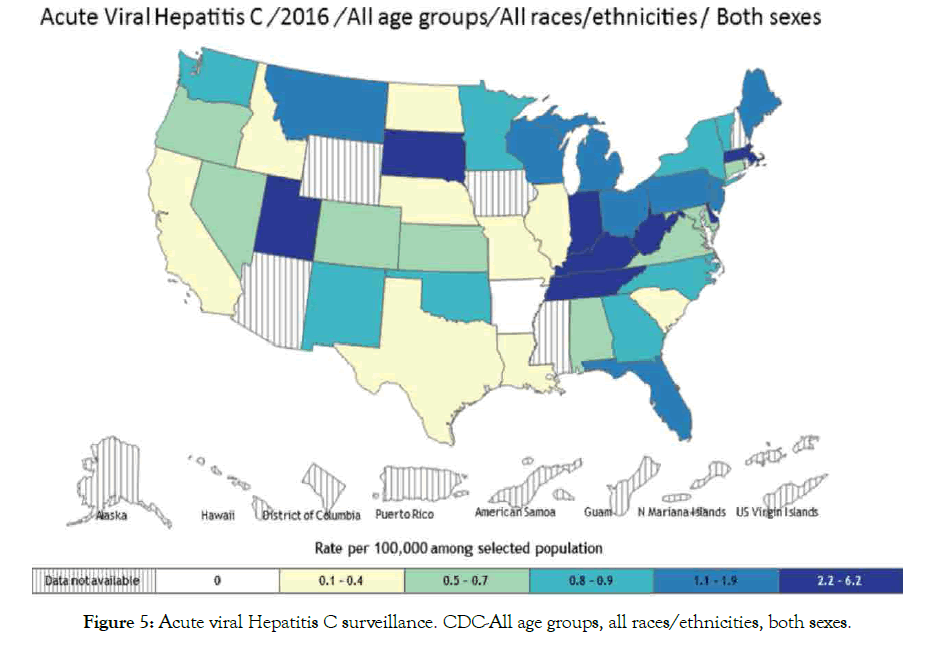
Figure 5: Acute viral Hepatitis C surveillance. CDC-All age groups, all races/ethnicities, both sexes.
Alcoholic, nonalcoholic and steatohepatitis: Two histologic patterns in NAFLD reported by Sanyal et al. are steatohepatitis and fatty liver disease although classified traditionally as alcoholic and nonalcoholic, of which nonalcoholic include both NAFL, NASH [4], study emphasized as an active NAFLD present with hepatic necroinflammation and fibrosis progression [107]. Obesity, lipid metabolism disorders, insulin resistance associated with NAFLD and other risk factors [108]. Worldwide estimates NAFLD prevalence at 25.24% high in Middle East, South America and low in Africa [24,109]. Nonalcoholic fatty liver disease prevalence ranked between 19-46% [110,111], which has continued to increase [112]. In addition, between 10 million people estimated coexist with NAFLD and diabetes in United States of note clinical variables can be employ as a guide in referral [113]. Metabolically, a syndrome associates insulin resistance, T2D, obesity as major risk factor [8,109], moreover hyperlipidemia, high blood pressure, diabetes and metabolic syndrome however inclusive [109]. Study established that low T1D and high in T2D prevalence in NAFLD could be latent autoimmune diabetes [114]. Furthermore, childhood and adolescence overweight associated with NAFLD, also in lean patient of which environmental factors such as high fructose, high fat diet and genetic term the cause. Endocrine disorder and drugrelated cause also reported [24]. PNPLA3, MBOAT7 and TMC4 have been associated with NAFLD [24], TM6SF2 identified in another studies [24,107]. Elevated level of TRIM24 reported in HCC of note originally termed transcription intermediate factor α TIF1α [115].
Disorder and fat accumulation in liver can generate to NAFL (usually present with fatty liver) and progressive subtype of NAFLD called nonalcoholic steatohepatitis NASH (present with fatty liver and inflammation, scarring leads to cirrhosis, hepatocellular carcinoma HCC and death associated with older age). The liver cells being replaced by scar tissue as a result of severe damage unable to function well determine by liver biopsy in NAFLD patients which may progress to cirrhosis [109,116,117], noting that NAFL cause globally liver disease. Nonalcoholic steatohepatitis quite significant using a diagnosis of NAFLD to establish prevalence and progression creates challenges for screening not keeping out origin as alcohol intake, hepatitis for fatty liver [109]. Studies further review factor contribute to NASH development like environmental and genetic factors of gene association with adiponutrin/PNPLA3 genotype associated with NAFLD progression to necroinflammation and fibrosis [24,118], and TM6SF2 through increased hepatic triglyceride [24,107,119]. SNP array development technologies for profiling common variant made it easier for GWAs approach in identifying risk genetic variant associated with ALD and NAFLD probable better than candidate gene approach [6]. Farnesoid X nuclear receptor (agonist OCA) showed efficacy, improve biochemical and histological feature in NASH, noting pruritus on 23% patient and overall increase in some cholesterol levels suggest be heated by positive findings [8]. Histological features for NAFLD include steatosis, necroinflammation and fibrosis, with tendency to progress from NASH to cirrhosis and other liverrelated complications especially NAFLD low activity scores [107]. Obeticholic acid, a semi-synthetic BA analogue (CDCA) and natural FXR and NR agonist [120]. Study establish that alkaline phosphatase and bilirubin level of 5-10 mg daily doses of obeticholic acid decrease in primary biliary cholangitis patients at phase 3 trial (56% 5-10 mg, 68% 10 mg, 38% placebo), for 12 months resulted with pruritus [80]. However, daily doses (50 mg) OCA for 2 weeks at phase 2 trial showed greater decrease and pruritus greatly severe, UDCA has remain approved treatment for PBC [72]. Study find that in overall patients 27% progressed to fibrosis, 48% with static disease and 25% with fibrosis regression [107], consistent with another study showing that in overall patients 42% progressed to fibrosis, no change seen in 40%, 18% regressed to fibrosis [121]. The grading and staging of NASH are not been validated, although grading classified from 1-4 base on fibrosis degree [4]. NASH presence, stage of fibrosis on prognosis showed no increased liverspecific morbidity and mortality, which can use in individual patient counseling and generally (Figure 6) [122].
Figure 6: Progression of NAFLD–Cirrhosis.
Hepatocellular carcinoma: Primary liver disease associate with death in cirrhosis from chronic liver inflammation development have been link to hepatitis BC infection. The incidence on liver diseases like cirrhosis follows likely pattern of variation with same etiology factors [123], that account between 70% to 90% of primary liver diseases [124], occur in 1-6% of PBC patients annually. The risk factors include male gender [124,125], old age, advanced histological stage, portal hypertension [126], and biochemical nonresponses to UDCA [127-129]. Other risk factors recognized are hepatitis B, C, alcoholic liver disease, NASH, blood transfusion [128], metabolic syndrome [124], and adenomatous hyperplasia as a precursor of HCC presented with malignancy disorder [130], lymphocytic piecemeal necrosis also associated with HCC. DNA methylation is important in development of tumor cells, it is necessary to ascertain when demethylation and hypomethylation occur during tumorigenesis. Study had it that hypomethylation occurs in HCC, which confirm of DNA from chemically induced hepatocarcinomas compared to DNA of normal rat liver, that showed an extensive demethylation of DNA in tumor progression [131]. More so, the degree of methylation nonmetastatic tumor is same as in normal prostate DNA, age not ascertain [132] Study showed higher degree of genomic wide hypomethylation associated with late grade in HCC which progresses in tumor size throughout life time [133], transposable element in HCC activated via genome-wide hypomethylation not ascertain. Reduction in genome scale DNA methylation and tend to distinct global DNA methylation level which can be used as a predictor in severe or complicated cancer types, [134] reveals common cancer hallmarks and specific DNA methylation pattern [135]. Alcohol consumption also accounts one of factors at high risk associated with HCC development in liver cancer [124], comparing HCC patients with NASH at 2.6% and NASH with cirrhosis at 4.0% per annual [117]. Patient with advance stage of primary biliary cirrhosis are at great risk for hepatocellular carcinoma development thereby requires surveillance, using known clinical variables which includes male sex, advanced age, portal hypertension, previous blood transfusion and stage of histology [128].
The incidence of specific (hepatobiliary) malignancy in PBC patient is at increased risk of developing cancer especially an advanced histologic stage at increased frequency [136], with evidence of low increase in overall cancer except liver cancer showing high prevalence and mortality in PBC patients [137]. Adding that HCV super infection may play roles in HCC development and history of cigarette smoking indicated on logistic regression analysis [138], more so, development HCC in PBC patient associate with 12 months biochemical nonresponses [127]. Study suggests of long term aggressive therapy in PBC patients may prevent HCC development [129]. Some strategies suggested proven effective and economic in preventive approaches to reduce risk of cancer incidence than treating an already advanced development of cancer , thus prescreen cancer patients at high risk more likely to benefit from preventive strategy [139]. Of note inflammation can impacts every single step of cancer development and progression even response to therapy. Whereas cytokines as a prognostic biomarkers contribute to a tumor microenvironment that permissive to cancer progression and related to drug resistance, which can disrupt long term success of treatment of cancer [140]. Anti-immune checkpoint inhibitors developed to avoid immunopathological diseases has been define for over active immune response and unsuitable reaction [141]. Active anti-tumor immune responses and modulators of specific genes have been associated successful oncotherapy either by presenting tumor antigen to activate or reactivate the adaptive antitumor immune responses [142]. The pro-inflammatory cytokines induction and activation of antigen-presenting cells associated with tumor ablation, thus immune defense that are targeted by immunotherapy showed effect in treatment[143]. Study of Yue et al. showed that immune system can educate cancer and suppress immune response and condition microenvironment provides interaction between HCC and immune system using patients-derived xenograft HCC-PDX technologies [144]. HSD17B13 rs7261567: TA associate reduced risk ALD, NAFLD, alcohol and nonalcoholic cirrhosis and lower odds of HCC (p꞊0.047) [94]. Study showed half of patients with advanced HCC have high levels of AFP that shows poor prognostic indicator and high rate recurrence in HCC patients with liver diseases. Although an advanced combine treatment using systemic therapy such as nivolumab and durvalumab expected to be more efficacious than single agents which absorption will be, oblige by safety and tolerance [7].
The current mechanism
Over last few years, identification of genetic diversity using various genomic analysis platforms such as SNP and aCGH. Of which CNV being at high-resolution microarray platforms to identify gene associated with various diseases in human. Genetic alteration and gene expression profiling analysis have provided information on the gene involve in various liver cancer [39]. As a result, various genes identified to associate in liver cancer such as tripartite motif-containing 24, 28 and 33 protein [21,145]. The DNA alteration of copy number variations involved has been identified in various human diseases and evolution [146], which are been used in diagnosis of complex human diseases [147]. Among CNV-driven genes identified, 8 of known cancer-related TFs which interact with 21CNV-driven gene, trim28 were included [145]. Identifying signaling pathways in liver carcinogenesis will provides new molecular target for therapy, such signaling pathways includes Hedgehog, Wnt-β-Catenin, tyrosine kinase receptorrelated pathways and propose becoming current molecular-based target for HCC treatment [39]. Of note the elevated nuclear β-catenin correlate with transcriptional target gene expression regulated by alternate signaling pathways in HCC that play roles in tumor progression and cell adhesion [39,148]. More so, increase retinoic acid signaling, activate single allele RARA and correlate with carcinogenic process which was able to completely restored suppressed hepatocarcinogenesis and expression of RA target genes at normal rate of hepatocytes proliferation [21,29]. Noteworthily, in a combinatorial and signal dependent way, KAP1/MyoD/Mef2D axis identified at transcriptional regulatory system [48].
The dysregulation of immune responses are associated with pathophysiology of various diseases incudes liver cancer. Study established link between inflammation, cancer development and progression as promoter [149,150], noting that NF-KB transcription factor serves as mediator of inflammatory process, play roles in innate and adaptive immune response [149,151]. Activation of nuclear factor (kappa B) in cancer diseases was associated in genetic alteration [149]. Whereas, proposed factors influencing signal-transduction mechanism in cancer that affect host through PRRs, regulate immune responses (innate and adaptive) and enzymes control expression gene encoding cytokines, chemokine [152], belonging to TLR family [153]. NF-KB either promotes or inhibits carcinogenesis on which inflammation and infection can enhance cancer development through signal-transduction mechanism influence in cancer surveillance and malignancy. Not with standing, NF-KB as a transcriptional regulator and seems to be the most signaling pathways that activated by infection, inflammation that can promote tumor in chronic inflammation and infection [154]. But specifically, in knockout mice model of multidrug resistance protein 2 gene MDR2 that lack P-glycoprotein in the canalicular membrane which leads to bile acids and phospholipids accumulation resulted to inflammation and rather HCC [155], also in chemically induced liver cancer with diethylnitrosamine DEN (mice lacking IkappaB kinase beta IKKbeta) exhibited an increase hepatocarcinogenesis [156]. Study identified involvement of phosphatidylinositol pathway in PBC that interplay with innate and acquired immunity [85,157]. The AKT and ERK1/2 signaling pathway activated by CBAs via SIPR2 hepatocytes and cholangiocarcinoma cells [158]. Identified a kinetic-based regulatory mechanism that determine between methyl and sulfur when limitedly available, it is the rate of cytosolic Fenton reactions that dictate nucleotide and RNA synthesis rate in cancer cell division [134]. Of which, Bacteroides fragilis correlate with GUDCA and TUDCA elevated level via methionine and folate modification inhibition. Thus, study revealed its beneficial effect dependent on bile salt hydrolase inhibition activities in B. fragilis via Amp-activated protein kinase-independent mechanism (insulin resistance and glucose tolerance) [159]. Consistent with other study beneficial effect of fexeramine modulate gut microbiota that induced acetatifactor and bacteroides with increase in bile salt hydrolase BSH thereby produces LCD from CDCA and UDCA through 7α- dehydroxylase and 7β-dehydroxylase activity [160]. In a different way, FXR regulate host metabolism and shape the gut microbiota by decrease FXR agonist BAs or increase FXR antagonist BAs in FXR signaling by large mass of BAs, importantly a positive feedback mechanism for regulating BA synthesis revealed [161]. Remarkably, study has reported mechanism that uses bile acids from gut microbiome as link to regulate chemokine which controls hepatic NKT cell accumulation, tumor growth inhibition (Figure 7) [162].
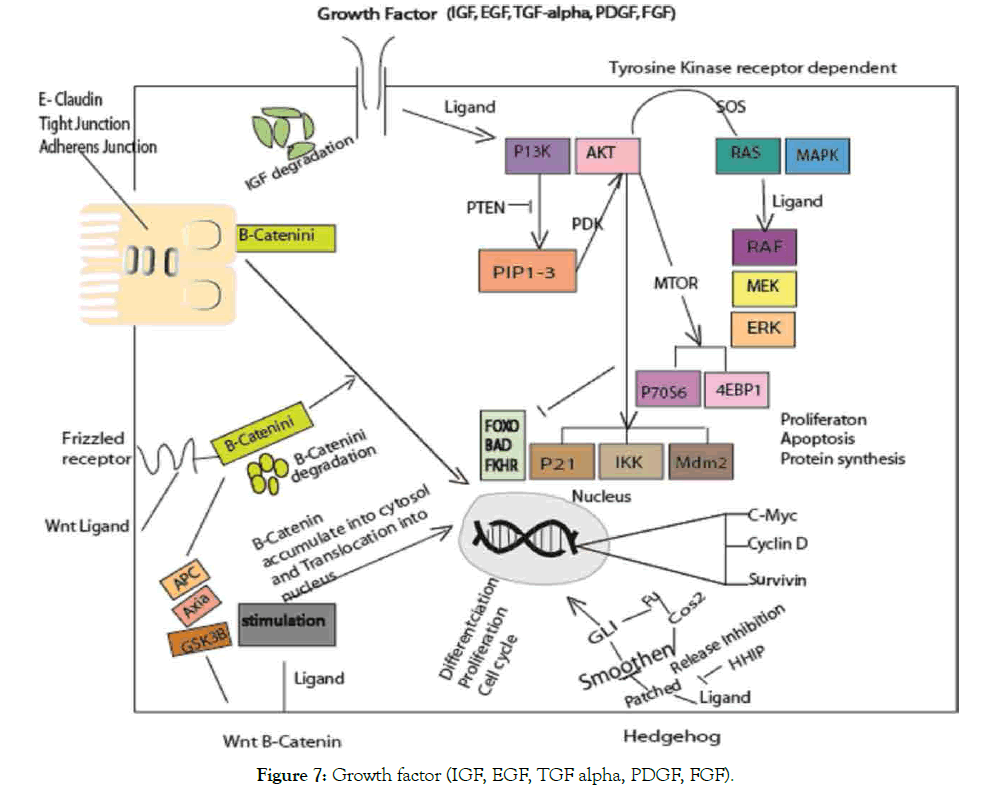
Figure 7: Growth factor (IGF, EGF, TGF alpha, PDGF, FGF).
Growth factors involve in regulation of biological processes. The different pathways involved in cancer. RAS/MAPK, PBK/ AKT associated with cancer both in cellular and intercellular differentiation. WNT-β-catenin and Hedgehog pathways disruption play roles in stem cell apoptosis. Translocation of Β-catenin regulate c-myc, cyclin-D and survivin from cell membrane to nucleus, of note act as coactivator of β-catenin enhancer associated with E-cadherin in the family transcription TCF/LEF [163]. At adherens junction, β-catenin associate with E-cadherin thereby link to actin cytoskeleton. In WNT pathway, β-catenin proposed degradation by phosphorylation, of which mutation will allow its escape to nucleus. Thus, enhanced target genes transcription that leads to cancer via different ways (inactivation or alteration) [164]. In hedgehog, blockade of PTC and SMO effect occur upon HH ligand that leads to the GLi to target gene. Frizzled activation via WNT pathway block β-catenin phosphorylation that leads to degradation of GSK-2β, APC and akin scaffold protein Akin. In TKR-dependent pathways, ligand bind to a growth factor triggers tyrosine kinase activity and transduction initiation. RAS/MAPK and ERK gene expression enhance proliferation. The two different mechanisms involved via MTOR induction through 5ʹTOPdependent and CAP-dependent (p70S6 and 4EBP1).
TRIM cofactors genes is the new target of the liver cancer
Genome-wide association studies identify sequence variants that associates increase risk in liver disease. TRIM24, TRIM28, TRIM33 were identified in liver cancer, thus showed much larger complex between TRIM24 and TRIM33 than TRIM28 and TRIM33 [21]. In line with identified alteration in gene expression associated with tumor liver formation and loss of function in gene mutation TRIM24 and this expression level were significantly altered in tumor of 55 genes resulting in 2.1- to 392 fold upregulated and 3- to 10 fold downregulated [29]. In addition, interaction between potent DNA-binding-dependent transcription repression domains TIF1 β and not TIF1 α and γ, krȕppel-associated box domains containing zinc finger protein is corepressor [165]. TRIM family member interactions interface between various receptors such as AF-2 activating domain of nuclear receptor TIF1, mSUG1 depends on AF-2 fusion and nature of receptor. The activation factor 2 of NR mediated transcriptional activity via different mechanism [166]. TRIM24 target endogenous p53 degradation confirmed negative pathway which regulates p53 drosophila and serves as therapeutic target thereby restore tumor suppression [167]. TRIM33 acts in different pathways of TGF β receptor are by phosphorylate drosophila mothers against decapentaplegic protein SMADD 2/3 as cofactors and as a monoubiquitin ligase for SMADD 4 [168,169]. TRIM24 genes identified to deregulate germ line mutant in liver [29]. Importantly, inactivation of TRIM24 tumorigenic effect was associated with deregulation of RA signaling in HCC, acting as a repressor to retinoic acid RA signaling and tumorigenesis in hepatocytes [29,38]. A subset of endogenous KRAB/ZNFs upregulate pluripotent stem cell [26]. HCC developments were significantly suppressed which observed in single retinoic acid deletion of alpha-receptor in TRIM24 null mice [29]. Notably, RARA expresses alteration of oncogenic activities correlate with dysregulation of retinoic acid signaling pathways in the postnatal developing liver [29]. Retinoic Acids (RA) a metabolite of vitamin A, it controls function of vitamin A that requires for growth, development and exert it biological function through binding and activating nuclear RA receptors RAR such as RARα, β, γ isotypes of which its impairment have been associated in various cancers [38]. Whereas, chemically induced transcription to prevent liver cancer by diethylnitrosamine DEN that accelerated in liver development, confirming TRIM24 as a tumor suppressor [38], recognized to regulate diverse cellular activity such as DNA repair, mitogenic signaling, protein ubiquitination and degradation [170]. TRIM33 function via inhibition of TGF-β signaling and execute their function in different way such as by inducing apoptosis in HCC cells and cytostasis [171,172], furthermore, induced different signal in other mediate liver tumor progression and invasion [173]. However, treatment improves apoptosis induce by different pro-apoptotic stimuli in TGF-β, highly express in liver tumor and cytokines [174]. Genetic alteration has been report of different pathways that implicated in liver cancer, revealed by genome wide allele-type studies and molecular cytogenetic analysis that inactivate p53 tumor suppressor gene associate to aflatoxin B1 exposure and HBV infection of which activation of cyclin D1 disrupt of Rb pathway involve in liver tumorigenesis [175]. Wnt-β-catenin pathway activated via CTNNB1/β-catenin mutation [40,41], AXIN1 also RB1 inactivation and IGF2R pathways via RB1, P16, IGF2Rs [40,175], described the structural and functional alteration of hepatocarcinogenesis that signaling pathways has been implicated [39]. It is essential to know that TRIM 33 and TRIM24 can interact via RBCC domain of KAP-1 using purified component, showed that KAP-1-RBCC oligomerization necessary for KRAB domain binding stating three of TRIM component act together in KRAB recognition [32,176,177]. However, the existence of TRIM28 and TRIM33 in same complex with TRIM24 was shown, as TRIM33 and TRIM24 most abundant and TRIM 28 least abundant [21,177]. Suggest that TRIM24 repress RA while TRIM33 modulate TGF-β signaling pathway [21].
TRIM24 overexpression were associated with HCC onset and progression, whereas knockdown TRIM24 showed an increases the p53, Bax, Caspase-8 protein levels, on the hand decreases Cyclin D1, CDK4, Bcl-2, survivin suggesting role TRIM24 play HCC pathogenesis [115]. TRIM24 can promote progression of tumor, recent study confirmed association with increased levels of TRIM24 protein in breast cancer and progression of prostate cancer due to overexpression of TRIM24 [178], other study report that poor prognosis and metastasis found in HCC may have been promoted by TRIM33 [179]. Moreover, TRIM24 interact with TRIM28, TRIM33 formed regulatory complex that suppressed in HCC and TRIM24-/- develop tumor in the liver [21,38]. Thus, genetics of knockout mice showed an increase of HCC that are significant implicated RARA in liver specific cancer [38]. TRIM24 knockout expression of apoptosis-related proteins observed in Caspase-8, Bcl-2, Bax, p53, Survivin, that upregulated in Bax, Caspase-8, p53 TRIM24 depletion, downregulated Survivin, Bcl-2 [21]. Consistent in the role TRIM24 play in proliferation and apoptosis as directly ubiquitinated p53 negatively regulate protein levels in breast cancer [167]. Correspondingly, in suppression of cell apoptosis, promote differentiation of hESCs via ubiquitination p53 [180].
Participation of p53 (tumor suppressor) mutation in different biological processes was established, of note showed its important role in HCC development [181]. Inhibition effect of TRIM member family significantly promoted in HCC development and progression through regulation of p53 pathway downstream signals [182], which inhibited colony formation in vivo by deceased tumor growth [182,183]. Remarkably, liver specific deletion of p53 in cancer was confirmed (loss of p53) independent of transforming growth factor beta TGF-β [184]. Additionally, TRIM58 function as tumor-suppressor gene via inhibiting cancer cell invasion through EMT, MMP activation, observed in highly overexpressed inhibition of colorectal cancer CRC cell invasion and lesser effect on proliferation and migration [185]. Furthermore, study showed significant in reduction of expression which down regulated in liver malignancy DRLM at deleted region termed class 11 tumor suppressors in HCC [43,186]. It has been reported also, that PNPLA3 was ending, variant rs738409, p.I148M associate with hepatic glyceride level, cirrhosis, NASH increase [90,187,188], and TM6SF2 encoding membrane 6 superfamily member 2 associate with NAFLD [91,119]. Abul-husn et al. identified new gene variants includes SLG39A12 encoding solute carrier family 39 member 12, GOT1 and GPT, both encode ALT and AST respectively. These have previously identified by other studies, also variant rs7261567: TA (HSD17B13) encode hepatic lipid droplet protein, associates with ALT and AST level reduction (p꞊4.2×10-12 and p꞊6.2×10-10) in NASH [94]. Another studies reported identification of PNPLA3 [90], TM6SF2 [91], SAMM50 [92], ERLIN1 association in liver disease [93].
Trim cofactors gene can regulate liver cancer via gut microbiome
Gut bacteria can regulate liver immune responses through both primary and metastasis cancer of which gut microbiome composition affected by different factors (genetics) stated below. First, let us look into gut as a microbial inhabitant that interacts with immune system influence innate and adaptive immune function. Emerging studies referred gut among the major sites of microbial inhabitant in human body, it harbor diverse microbes play roles in well-being of their host. The diversity of human microbiome composition and microbes in healthy and disease is complex [189]. The human gut microbiome composition can affect by different factors like genetic, environment such as dietary, drugs and intestinal motility [190] and physiology [191]. Genetic susceptible to disease influence on the gut microbiome varies among human distinct in genus and species [190]. The individual connection and human associated inhabitants between microbes [192], immune function [193] and metabolite [194], which influence the development of microbes starting from birth (infant) to disease state [195-197]. These have established by different experimental, computational tool and technologies that has stored in database by different research project [13,198,199]. Although mechanism are less known but the early interaction of immune system to commensal believed to occur through the passage of birth canal and colostrum of which certain factor in breast milk like immunoglobulin A, immune cells-cytokines, metabolites living microbes in breast milk involved [157,196,200]. However, association of specific diseases to the taxa should be intense [190]. Microbiota diversity of uncharacterized gene and species based on large scale microbiome profiling projects [198,199,201-203], and rare variant of little effect that difficult to identify by genetic means [13], but systematic identification using high-throughput sequencing and human genome [204]. Emphasizing on privacy protection based on the control or handling of microbiome nature of these high resolutions tracking the microbial variables in future is indeed terrifying [205]. The human genetic variant studies associated with different human diseases [6,205], have established such as in obesity [206-208]. Asthma [209-211], inflammatory bowel diseases [212], hypertension, [213] diabetes [214,215] alcoholic liver disease [216], Parkinson [217], and liver [218] through mRNA and regulatory sequence of difference in translational variability [217,219]. The microbiome, genetic susceptibility, epigenetics regulation, environmental factors involve in complex interaction in development and progression of diseases. Though genetic susceptibility usually demand as a basic etiology for disease condition, in the case of fibrosis with a classic Mendelian inheritance of which microbiome play roles in but might not be the only cause such as in Parkinson [217] and IBD [220].
The gut microbiome has intimate interaction with host immune system can influence by adaptive and innate immune functions [221], which include neutrophils, DC, NKT, development of Tcell subtypes associate to a particular microbiome [222], of which adaptive immune response involve in B and T cell activation. Interaction between gut, liver, immune system metabolisms thus showed microbiome involvement. Study identified gram negative bacterial (LPS) as a triggering factors situated at blood stream from gut at the early development of metabolic endotoxemia diseases of which insulin resistance associate with a low-grade inflammation [223]. The gut wall bacterial contain molecule LPS and when altered it becomes leaky whereby LPS goes into the blood stream, excessively dysregulates the inflammatory tone and triggers glucose metabolism. Noteworthily, gut microbiome function and bacterial composition change associated with dysbiosis result in an increase of oxidative stress thereby motivate a chronic inflammatory response [224], indicated that inflammatory response has connection between immune system and gut microbiome. Study identify organisms that have ability to exacerbate inflammation with a reduction among members of its microbial community known to produce short chain fatty acid [225], whereas gut epithelium deficiency fatty acid: butyrate acids have ability to raise an inflammatory responses [226].
The role diet and chemical play in disease management need to put under consideration. The metabolism of microbiome can modified by the host genetic, environmental factor based on dietary consumption and most changes are reversible although microbiota abundance depend on diet history [206,227-229], Shaping individual microbiome associated with complex polygenic traits that affect microbiota composition in three ways [228], and change in BAs. BA increase glucose uptake was confirmed and cold acclimation have effect on skeletal muscle insulin sensitivity of T2DM patients which lead to glucose transporter type 4 GLU4 enhancement [230], GLU4 allow facilitated diffusion of glucose down into muscle and fats cells which phosphorylated by glucokinase in the liver. Cold induction of thermogenesis is a mechanism to maintain body temperature in cold exposure in animal that has gained interest in BAT discovery. In addition, cold exposure mice reduced obesity, improve insulin sensitivity, energy expenditure [231], promote thermogenesis cold exposed proposes beneficial metabolic effect that mediated direct by BAs via TGRS or indirectly by changes in gut microbiome composition via alternative pathway [232]. The amount of energy expenditure activated by dietary induced thermogenesis DIT produces heat by brown adipose tissue BAT in cold exposure, BAT induction increases energy expenditure which associates with lean phenotype mice also mediate BA metabolism via FXR and AMPK signaling and bring about changes in gut microbiota [233]. Inflammation signaling and glucose metabolism modify by antibiotic induce in gut microbiota [234], and in early life murine metabolic homeostasis alteration [235], growth and energy homeostasis [236].
TRIM family proteins established its role in innate immunity, differentiation, migration, proliferation, apoptosis, tumor development and progression [237,238]. Importantly, it is involved in biological processes [237,239-241] TRIM24 (potent liver specific tumor suppressor) family proteins confirmed to take part in p53 regulation while TRIM28 in p53 inactivation of note are largely expresses in cancer [238]. Study identified role of TRIM28 (KAP1) as a cofactor and regulator of myoblast differentiation MyoD (controller of myogenesis) function of which phosphorylation of KAP1 were mediated by MSK1 thereby releases corepressor from the scaffold [48], thus involved in recognition and repair of DNA by serine 473 and 824 (cellular DNA damage response) [34]. TIF1s interact with transcriptional silencing domain of drosophila (KAPI), of which TIF1 gene family involved and influence in the initiation of transcription [36]. Likewise trim28 interacts with KOX1/ZNF10 that represses transcription [242].
The role of enzymatic reactions by BAs synthesized in liver via different BA synthesis pathways, encode CYP7B1 and CYP27A1 gene followed by 25 hydroxycholesterol 7-α hydroxylase gene [243]. Worthmann et al. identified CYP7B1 induction as a key BA gene increases synthesis and energy expenditure [244], which BAs identify as an important metabolic effector based on BAT activation cholesterol and energy metabolism by the host. The both conjugated with taurine and glycine, activate the transcription factor and nuclear hormone factor FXR via fibroblast growth factor FGF 15 and 19, thereby generate endocrine negative feedback signal to the liver and other effect such as glucose metabolism in T2DM, fatty acids and cholesterol [245]. The conversion of hydrophobic CDCA to hydrophilic differs between rodents and human in BA metabolism [246]. Study observes lower hepatic CYP7B1 gene expression in obese and T2D patient, attribution to the CYP8B1 induction, inhibition of CYP8B1 knockout mice altered by BA composition repress hepatic de novo lipogenesis DNL and gut microbiota homeostasis, DNL that account between 10-30% hepatic fat accumulation of triglyceride in the liver of NASH and NAFLD [247,248]. Although, without conclusion to the combination effect of BA synthesis on the microbiome composition of both human and mice connection to BAT activation. The study of combination effect on microbiome composition suggested that BAT might have caused diversion in BA synthesis, observed in both mice and human [246]. In identification of BA, study indicated location of increase brown adipose tissue iBAT that upregulated energy expenditure, BA activate by BAT in human [249], and activates FXR and TGRS to regulate thermogenesis [250], promote energy expenditure and inhibit FXR activity of AMPK. Contrast to other study that AMPK activated [251], therefore increases expression of phosphorylated AMPK in the liver [252]. For instance, in absence of bile acids receptors FXR liver tumors developed in knockout FXR mice, indicating roles FXR play in BA regulation [253,254].
More so the activation of FXR in the liver controlled BA synthesis via negative feedback by reduction of secondary BA muricholic acid MCA (FXR antagonist) [255], also in gallbladder [256] of which state that gut microbiota play roles in BA metabolisms and regulation in liver [255].
Bile acid produces in liver from cholesterol, secrete by hepatocyte into bile canaliculi stored in gall bladder. Bile is being flow to duodenum after ingestion of food, help in solubility and digestion of lipid which absorb by passive diffusion, transport from terminal ileum back to liver via portal vein [257-259]. Bile involve in development of liver cancer, help to break down fats. Gut microbiome use BA shape liver immunity. The bile that secretes in liver altered by intestinal bacterial overgrowth, systemic infection which obstruct bile flow promotes in gut microbiota [260], thus leads to conversion of primary to secondary bile acids. Notable negative feedback regulation of BA synthesis disrupted mutant mice thereby cause alteration that leads to liver disease [261]. The metabolic pathways in which bile acid synthesize in liver, thus taurine and glycine are primary bile acid (CA - TCA, GCA and CDCA: TCDCA, GCDCA) conjugate as bile salts (7-α dehydroxylase) mediate cholesterol oxidation by the enzyme (cholesterol 7-α dehydroxylase) which downregulate cholic acid. The bile acids biosynthesis is downregulated by suppression of expressed CYP7A1 via c-Jun N-terminal kinase dependent pathway and upregulate by a protein FGF-19 secrete by the ileal enterocyte and mediates sodium dependent BA transporter (apical) which uptakes across ileal enterocytes [258]. In the intestine, bacteria transform this BA into secondary BA (deoxycholic ac DCA, LCA) that absorb into blood stream, returned to liver via enterohepatic circulation and finally enter liver acinus [159,262]. Cholangiocarcinoma have been significantly linked to liver diseases, thus evidence of an increase found in cirrhosis patient and associates to intrahepatic cholangiocarcinogenesis [123,263]. JTE-013, a potent selective antagonist S1PR2 bind to human, rats receptor, inhibited TCA, SIP-induce ERK1/2 and AKT activation in cholangiocyte, that reverses inhibition of SIP on invasion and migration of B16 melanoma cells. This study reported that JTE-013 specific shRNA of S1PR2 and chemical inhibitor of ERK1/2 and AKT inhibited TCA and SIP cell proliferation and migration in cholangiocytes mice [264].
Bile acid is the key regulator of glucose, lipid metabolism and energy homeostasis via membrane and nuclear hormone receptors by FXR (OCA) that have tested in primary biliary cirrhosis patient who does not respond to ursodeoxycholic acid [258,265]. Indeed FXR have molecular link between BA and plasma lipid play role in control of triglycerides [266,267], and cholesterol metabolism [268]. Remarkably, patients responded to UDCA therapy showed normalize survival rate when given at early stage of PBC [269]. Importantly, farnesoid X receptor agonist (OCA) decreased hepatic steatosis, insulin sensitivity in T2D and NAFLD [120,257,270]. In addition, inhibition of lipogenesis due to FXR activation, function as the suppression of CYP7A1 in human elevated level of deconjugated TUDCA and GUDCA that suppressed by decrease of Bacteroides fragilis bile salt hydrolase BSH [159]. Study report that FX nuclear receptors signaling affect lipid metabolism in the liver, which can change with FX nuclear receptors ligands [8]. As ligands for GPCRs- TGR5 (GPBAR1 gene), membrane bile acid receptor locates on chromosome position 2q35 in humans, encoding 330 amino acids with 993 base pairs open reading frame, TGR5 mRNA expression detected at high level in various organs like small intestine, liver, stomach, lungs [257,271]. Given that change in protein expression involve in production, secretion and recirculation of BA can alter BA signaling, transport and modulate expression and activity [159,257]. Noting that the nuclear receptors FXR, transcription factor modulate production and BAs metabolism in liver. For instance, the growth of Bacteroides fragilis inhibited by metformin via folate modification, methionine metabolism and intestinal FXR signaling via gut microbiota modulation [159], In addition, activation of FXR prevented in vivo induced synthesis oxidative stress enzymes, proteinuria and glomerulosclerosis in diabetic kidney disease using FXR modulation [270]. TGR5 is not for bile alone but various selective synthetic agonists such as 6EMCA, 4-benzofuranyloxynicotinamide derivative to regulate different signaling pathway like NF-kB [272,273]. On the other hand AKT, ERK metabolic regulator involve in glucose metabolism, bile acid and energy homeostasis, even in inflammatory response, cancer and liver regeneration [274,275], which takes part in physiology of liver and gall bladder [257]. Importantly, TGR5 activation of bile acids increase GLP1 and insulin secretion, in line with, FXR inhibition through GUDCA elevates serum active GLP1 level (Figure 8) [159].
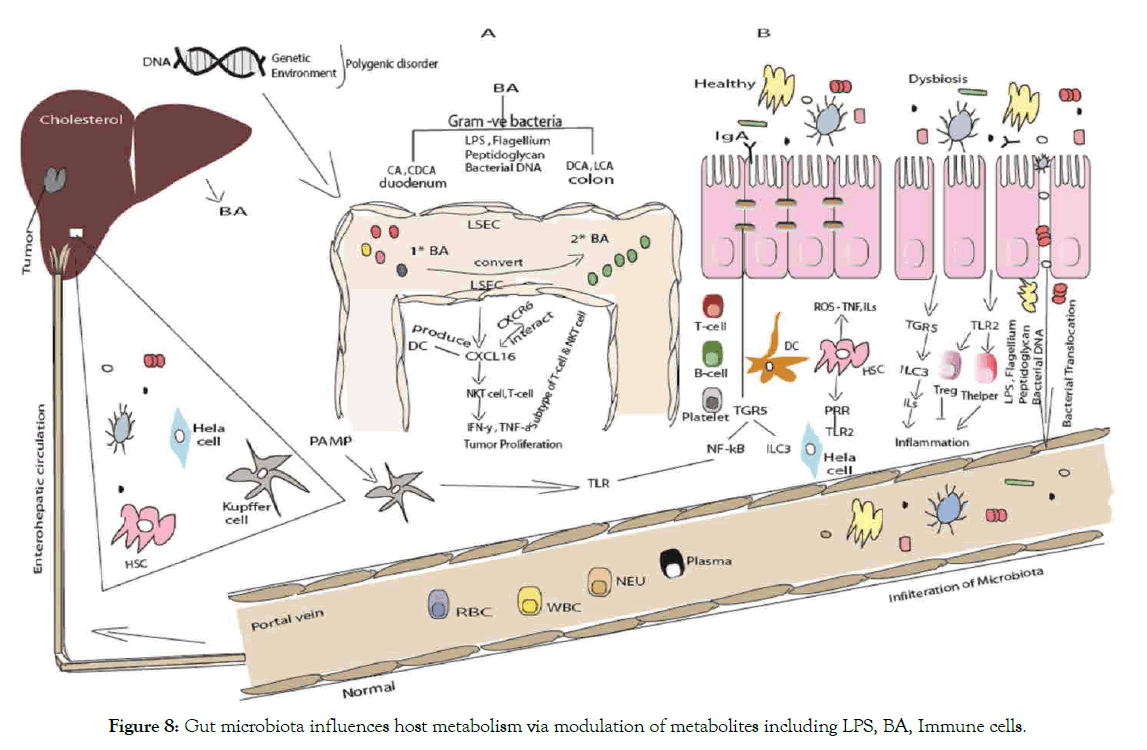
Figure 8: Gut microbiota influences host metabolism via modulation of metabolites including LPS, BA, Immune cells.
(A) Liver exposed to gut bacterial metabolites through blood via portal vein. Liver produce cholesterol through hepatocytes in the liver synthesis and transform primary into secondary BAs by enzyme hydrolase, 7α-/β-dehydration in intestine [276]. Bacteria in the gut mediate the metabolism of bile acid and regulate liver cancer through NKT-cells. Microbiota modulates the expression of several enzymes via bile salt hydrolase. Hepatocytes produced primary bile acids that transported into the duodenum. Some bile acids reabsorbed and other remaining convert from primary bile acid to secondary bile acid as result intestinal bacterial action in the colon. Secondary bile acid inhibit the activation of Liver sinusoidal endothelial cell LSEC while primary bile acid promote protein chemokine ligand 16 CXCL16 production by LSEC, an increase expression of CXCL16 will recruit NKT cells [162]. Of note that NKT cells produce IFN-γ, ILs thus has antitumor effect. After travel through duodenum to colon, most of the BA will recycled. It will be absorb at terminal ileum and recycled through enterohepatic circulation. Primary BA stimulates the expression of CXCL16 that interact with the CXCR6 receptor. Endothelial barrier consist of LSEC that regulate the hepatic immune homeostasis. Nuclear receptors and membrane bound receptors regulate metabolic immune related processes [277].
(B) Epithelial barrier bound across tight junction. The innate and adaptive immune cells maintain homeostasis at steady state. Alteration allows luminal microbiota to trigger unlimited inflammatory response. Factors contribute to disruption of tight junction are inflammatory cytokines secreted from activated immune cells in lamina propria. LPS translocation occurs through intercellular space to systemic circulation. LPS and bacterial reach the liver via portal vein and metabolize or acts as ligand of PRR such as TLR. Bacterial LPS acts as a triggering factor [223].
Trim cofactor genes can regulate liver cancer immune cells
Human body comprises of defense system that made up of entire organs and vessel system such as lymph vessel and individual cells and proteins. Immune system is system comprises specialized cells, organs that protects against organisms or invaders called antigens and respond immediately. They classified as innate and adaptive immune system [278], contain humoral, cellular component in each immune response that recruit, activate immune effector cells which are the resident immune cells (granulocytes: basophils, eosinophil, macrophages- recycling dead cells like RBC, fibroblasts, mast cells [279] releases inflammatory chemical like histamine, dendritic cells (antigen presenting cell APC). Natural killer cell (innate and adaptive) cause apoptosis do not release signal leads to immune activation and inflammation. Leucocytes like monocytes develop to macrophage and neutrophils that possess surface receptors as PRR (TLRs, NLRs) [280]. Of note loss of negative regulation of TLR signaling was associated with the pathogenesis of autoimmune diseases and inflammation as Atg16L1 negatively regulate TRIFdependent pathways cause casepase-1 activation [281].
Liver is exposing to pathogen or microbe-associated molecular patterns through PRR such as TLR and NLR. Immune responses (innate) are triggered by TLR play role in innate immune responses and other PRR by viral [17], fungal and bacterial infections. The loss of negative regulation of TLR signaling associated with autoimmune and inflammatory pathogenesis, thus play role in host defense and diseases [281,282]. The involvement of five adaptor protein by TLR signaling, SARM negatively regulate TRIF induce IFNβ for activation of interferon IRF3 family [283]. Thus bind to one -two classes of molecules such as PAMPs (β-glucan of fungi, endotoxin or LPS of gram-negative and LTA of gram positive bacteria) [284,285]. Of note, immune recognizes P/MAMPs via PRR as stated above. PAMP with some endogenous molecules play roles in facilitating adaptive immunity against infected microbes thereby activated innate immune via TLRs and PRR as an adjuvant [281]. Study identifies LPS and bacterial DNA as a prototype of P/MAMPs [286]. In established model of two strain mice (C57BL/10ScCr and C3H/HeJ) unresponsive to LPS highly resistant to LPS induced shock by TLR4-deficient mice [287]. The damage associated molecular pattern DAMPs [278,288] either repair, stop, invade or remove foreign particles and host debris by phagocytosis to the infection site through chemical mediators production like chemokine and cytokines [289], tumor necrosis factors TNF-α, IL-1, 6 and IFN-Y can lead to cancer when activated. Of note, involve in inflammatory changes that occur during obesity development thus affect adipose tissue inflammation [290]. It have been found that SIBO increase intestinal permeability, alter by colonic microbiota composition which releases pro-inflammatory cytokines LPS to reach liver via portal vein to systemic circulation [291]. The innate signaling regulation response also triggered by toll-like receptors TLR signaling lipopolysaccharide LPS-induced transcriptional regulation of inflammatory responses [280,289]. In addition, the innate immune cells can activated via PRR by immunogenic cell death ICD induce by injury, stress and chemotherapeutic agents in cancer cells [288]. As type 1 IFN production was impairment resulted in the loss of TLR signaling [292]. In recognition of microorganism by PRR triggers activation of antimicrobial defense, stimulates adaptive immune responses [293]. Disruption of intestinal barrier leading to increase permeability, which influences the bacterial metabolite and PAMPs/MAMPs that liver are expose to. Of note liver feeds back to the intestine through bile acid secretion and mediators. Immunoglobulin A produced by B-cells, of which it plays important function in host microbiota homeostasis regulation [294].
Adaptive immune system consist highly specialized system (T and B) cells [278,293]. The process that eliminate or prevent pathogenic growth, which are being triggered when pathogens invade innate immune system and generate a threshold level of an antigens and signals [281,293], it start to work after innate immune system is activated, dendritic cells thereby initiating antigen-specific immunity [295]. The mechanism of adaptive immune system activate via three pathways (classical, lectin and alternative) [279]. PRR interact both exogenous and endogenous ligand via APC integrating signal with cellular interaction to generate diverse responses [284]. This provide the body ability to recognize and remember by these specific pathogen (APC) such as dendritic cells, also by macrophage via phagocytosis which travel to lymph node- and bound MHC class II and MHC class I receptors APC. These correspond to dendritic cell macropinocytosis of antigens that identified [284,296]. The monocytes derived-dendritic cells MDDC as rottlerin inhibitor of macropinocytosis [297], presents to immature helper and cytotoxic T cell through binds to MHC class II (Thelper CD4+ cell with four subsets- Th1, Th2, Th17, Treg) or MHC class I (CTL). Resulted in upregulation of MHC II, cytokines, chemokine and T cells via antigen receptors [295], and T cells lymphocytes matures and proliferate, the helper T cells activate B cells (expresses in two ways) which will later proliferate and produce antibodies (immunoglobulin Ig- IgM, IgD, IgG, IgA, IgE) specific for antigen [298]. On the other hand cytotoxic T cells destroys pathogens presented by APCs, finally T and B cells (lymphocytes) that activated become memory cells (passive or active) as memory T and B cells. More so, over 50% B-cell from HCC tumor activates showed high reduction FcyRII [299], thus established inflammation liver disease CD8+ T cells accumulation. In correlation with tumor-derived CD8+ T- cells dysfunction that have ability for proinflammatory TNF-α, IFN-γ production, cytotoxic GzB and perforin [299]. Furthermore, T-cell induced mucosal damage by perforin via combined effect of different pathways of cytotoxicity like IFN-γ, TNF-α, perforin and Fas/FasL Fas ligand (play role in regulation of immune system and cancer progression) [300], noting that perforin contribute to apoptosis through perforin pores causing granzyme introduction to the target cells. Interestingly, study reveals tumor cells release autophagosome TRAP induces B cell differentiation in IL-10 produces B cells and suppress T- lymphocytes activity [301], ameliorate neutrophil apoptosis in cell lines and tumor cells through macropinocytosis via caspase-4 and reactive oxygen species ROS generation, thereby inhibited CD8+ and CD4+ T proliferation [302], which could be employ as immunotherapy. Suppressed tumor-induce immune reduction in cancer patients, consistently in other study ROS which produced by NADPH oxidase play role in neutrophil cell death mechanism, through pro-apoptotic Bcl-2 and caspase activates either by necrosis or apoptosis [303]. Recent study revealed that TRIM33 (lineage dependency) associated with two lineage- specific enhancer PU.1 transcription factor TF (PU.1 TF), which negatively antagonize its function by same element that being recruited [51]. Among B cell acute lymphoblastic leukemia BALL cell analysis of gene expression, Bim and Atp1b3 upregulated in TRIM33 reduction [51]. In line with study, that Bim play role in B-lymphocytes deletion and Bcl mediated apoptosis because of loss Bim, which inhibit the deletion of autoreactive B cell [304].
Adaptive immunity of IgA+ cells prevents cancer envelopment; suppress liver cytotoxin CD8+ T lymphocytes CTL activation in HCC and Mice [299,305]. Study of cancer immunoediting mechanism involves the use of combine immunotherapy that shaping tumor fate in three phases, increased therapeutic index [306], of which mechanism dependent of NK cells and IFNγ [307]. A combination of antitumor and CpG–a TLR9 agonist trigger activation of the systemic antitumor immune response remarkably shows a therapeutic memory response in mice expression of OX40 and CTLA-4 immune cells upon tumor recognition antigens that inhibited by tumor-specific Tregs via upregulation of TLR9 [308]. Furthermore, the combination of anti-OX40 AND CTLA-4 with TEA therapy enhanced better responses [309], which maintained a constant internal environment for homeostasis clearance mechanism of the host pattern recognition [284]. Studies identify human subset and type II NKT_TFH cell regulate B-cell contrary to gaucher disease, metabolic lipid disorders via glucosphingolipid mediated pathways and dysregulation of humoral immunity [310]. Type 1 IFN activation upregulate PD-1-PD-L1 signaling axis in tumor tissues by NK and T-cells indicates its beneficial therapy PD- 1-PD-L1 blockade [311]. Natural killer, NKT and Dendritic cells may be responsible for innate immune response limitation (impaired) and degeneration in HCV through various pathways [312], thereby generate diverse responses from exogenous and endogenous ligand by APC with cellular interaction responses [284]. IFN-γ can induce autophagy by different proteins, establishing its role in autophagosome formation [239]. TRIM24 negatively regulate tumor suppressor p53 levels established TRIM24-p53 links [167]. TRIM33 act as tumor suppressor in human CMML and mice thus play role in preventing the onset development of CMML [55]. Suggesting TIF1G regulate epigenetically tumor suppressor gene in hematopoietic cell. Studies reported relationship between TRIM family protein and autophagy. Based on recent studies that TRIM proteins interaction with p53 protein [313]. Another study reported autophagy-related Atg proteins involved in autophagosome formation [314]. In addition, p62 mediate Nrf2 activation because of ubiquitinated protein accumulation in damaged tissues [315]. Autophagy referred as a natural occurring cyto-protective mechanism degradation that play crucial roles in induction both preventive and therapy of healthy and tumors cells [316]. Thus, show alteration of HIV infection through the process of autophagy [317]. Those induce by chemotherapy, autophagy by starvation, growth factor deprivation and hypoxia [318]. Programmed cell death or apoptosis are two different ways by which cells respond to stress [319], that can inhibited by antibiotics, of which lgG (IgG1 and IgG3) is most used in cancer immunochemotherapy [298]. The approach described for microautophagy and macroautophagy of which inhibitors that affect microautophagy steps not impair delivery of peroxisome to vacuole via macroautophagy [320]. Besides autophagy identify as promising target in cancer therapy, of which Beclin1- silenced B16F10 cells that secret TRAP have little LC3- II that correlates with decrease ability to induced PD-L1 and IL-10 [321]. Accordingly, glucoseinduced micropexophagy in two distinct pathways [322], more so autophagy-related three different pathways can also induce Atg proteins: starvation-induced pathways, Cvt pathways, pexophagy (autophagic degradation pathways peroxisomes in yeast cells), all to core machinery for membrane formation [314,321]. Furthermore, autophagosome modulate effects of immune cell function and tumor progression [301]. On which autophagy a normal protective survival mechanism for cells that undergo different ways of stress and formations in three major ways [309], started with the formation of autophagosome thereby releases extracellular fluid by inhibition of proteasomes or protein oxidation increase or accumulation of misfolded proteins of vitamin E analog on stress induction [301]. A semi synthetic vitamin E derivative (α-TEA) induced tumor cells apoptosis and autophagy thereby improves cross presentation tumor antigen on lung immune and murine mammary tumor cells [309]. TRIM family protein directly or indirectly involved in regulation of autophagy, carcinogenesis and immunity, it regulate immune responses through innate immune signaling such as NFKB signaling and IFN signaling [313].
Tripartite motif containing proteins play broad range of function on inflammation [323]. Thus, exert its regulatory role in various biological processes like differentiation, growth, apoptosis, carcinogenesis and antiviral immunity [237-241]. These roles were observed of TRIM8 in inflammation, cancer regulation expressed pro-inflammatory cytokine and IFN related transcription factor [324]. Furthermore, negatively regulate the interferon promoter activity and in different way regulate the transcription of pro-inflammatory factors in overexpressed EcTRIM13 [325]. Investigations supported by some other studies showed those liver progenitor cells activated when mature hepatocyte compartment are damaged due to chronic inflammation or toxic injury [326]. For instance, of cells levels that respond to carcinogenesis in the hepatic lineage such as mature hepatocyte, ductular progenitor cell and putative periductular stem cell [327]. Of those, different cell signal involve in this process such as immune cell, growth factor [328,329], cytokines which are produced by various immune cells like macrophage, B and T lymphocytes, mast cell act through receptors. On the bases of cytokines network initiation through the binding of TNF-TNFR1type 1, activated NF-KB in NPC to produce IL-6, STAT3 activation in hepatocyte [330]. Given that, oval cells (hepatic stem cells) significantly unchanged impaired TNF type 2 knockout mice noting IL-6 reduction suggest TNF signaling contributed to this effect [329]. However, study emphasize that it originated from HSC of which migrate to liver and differentiate into hepatocyte [326,331]. Furthermore, oval cells expresses high level of Sca-1, CD35 and CD45 [331], Correspondingly supported by another study that the fusion between hepatocytes and transplanted hematopoietic cells possibly mechanism by which hepatocytes are being generated [332]. Progenitor cells said to be the progeny of stem cells and when isolated from certain organs depends on ability to differentiate into different TSP (plasticity) such as liver (portal space-bile duct cells and parenchyma-hepatocytes) [326]. Importantly, showed evidence of cytokines pathways activated between Kupffer cell and hepatocyte in liver regeneration and metabolic pathways [330]. More so, nuclear factor-kappa β [333], STAT3 [334], C/EBPβ [335], and transcription factor plays role in liver regeneration in sense that it can extend signal by activating various genes [336].
Tripartite motif protein family contain ring finger domain also involved in several of cellular functions. Study observed regulatory innate immune response through modulation of PRR signaling pathways [17]. Cells possess surface receptors as PRR which bind to PAMP like protein, lipoprotein, lipids, nucleic acids [292]. Moreover, various signaling pathway involve in NF-k B, AP-1, IRF protein activation, PAMP recognition by TLR dimers has been identified and characterized [281], besides its role in host defense was established [17]. Furthermore, TRIM family proteins directly interact with cellular proteins as a combined effect or single thereby modulate signaling pathways that triggers engagement of PRR. The expression pro-inflammatory responses (cytokine) affect IFNs (type 1 and 11) by downstream regulation, also promote adaptive immune responses [19].
NK and NKT cells
Liver is immunological organ that are circulate mainly by immune cells and expose to gut microbiota via portal vein. The liver diseases are usually associates with altered gut bacterial composition. Gut commensal bacterial is an important regulator of antitumor immunity, reported that NKT cells is a regulator of autoimmune responses [337]. According to one study that alteration of gut commensal bacterial induces liver selective antitumor effect thereby increases hepatic CXCR6+ NKT cell, NKT cells accumulation were regulate through expression of hepatic CXCL16, a ligand for CXCR6, as primary bile acids increases expression of CXCL16 while secondary bile acids reverses it [162].
Natural killer cell is an effector of innate immune system, serves as a natural regulator to adaptive immune responses [338]. They have features both innate and adaptive immunity that is subset of lymphocytes in peripheral blood (lipid), although do not expresses membrane receptors that distinguish B and T cell lineage, they lack antigen-binding receptor, memory and immunologic specificity [339]. NK cells produces in high amount interferon-gamma IFN-γ that are used to fight viral infections [104], consistently, elevated IFN-γ production upon antigen stimulation [162], that have ability to kill tumor cells in interactions between ligands of the tumor cells and various receptors on NK cells, releases cytotoxic granules of the NK cells which produces inflammatory cytokine [340]. NK cells express activating and inhibiting receptors. While activating receptors and associated with signal transducing molecules [341].
PRR pattern recognition receptors molecules interact with both endogenous and exogenous ligand by APC (innate- macrophage and dendritic cells) and able to activate adaptive immune response (T- and B-lymphocyte) which plays dual role in host defense and normal tissue function [284]. Showing self-structures on normal, impaired, stressed and transformed cell of immune recognition mediated by NK and γδT cells involve in restriction of non-classic MHC class IB molecules [284]. The role LrNK cell play in T-cell immunity regulation were established mechanism of immune tolerance in liver, report showed that inhibition of hepatic T-cell function by LrNK CELLS caused impaired viral clearance via PD-1- PD-LI axis [342]. However, impair T-cells response induces by PD-1- PD-L1 axis showed an increase during viral infection, of which PDL1 significantly expresses on LrNK cell and PD-1 receptor increases on T-cell indicating that PD-1 blockade lead to improvement in immune response [343]. The liver residents and hepatocytes have been established for expression of CD1d [344], study had it that cholangiocytes also expresses CD1d which present antigen to NKT cells which play role in the immune system during liver disease progression thereby restricted activation [345]. NK, NKT and dendritic cells may have been responsible for innate immune response limitation (impaired) and degeneration in HCV through various pathways [312], thereby generate diverse responses from exogenous and endogenous ligand by APC with cellular interaction responses [284].
T–cells
CD4+ CD8+ is two T-cell categories base on which protein present on cell surface. CD4+ T-cell having four major T-cell subsets TH1, TH2, TH17, Treg refer as T-helper cells. All have different functions as TH-2 known to coordinate immune response against extracellular pathogen, TH-17 for recruiting neutrophils, TH-1 for immune response coordinating against intercellular microbes such as bacterial. They also produce, secrete molecule that alert and activate immune cell such as bacteria-ingest macrophage. Regulating T-cell Tregs function as inhibitor to the activities of other T-cells and monitoring. Also, can prevent activation of adverse immune response; maintain tolerance against body’s own cell and antigen. CD8+ T-cell (CTLs) known to recognize virus-infected and tumor cell removal, also stages programmed cell death (apoptosis) [319]. T-helper cells activate B-ells in two ways, which will later proliferate and produce antibodies. Antibodies are unique and fall into these various categories of immunoglobulin IgM, IgD, IgG, IgA, IgE [298].
Immunologic effect of microbiota homeostasis stimulates immunoglobulin A production, downregulate pro-inflammatory cytokine, promote anti-inflammatory cytokine, induces regulatory T- cells [291]. Study has reported that pro-inflammatory cytokines (IFN-γ) induce loss of granule thereby causes paneth cells degranulation [346], Furthermore, butyrate induce differentiation of functional colonic Treg through intrinsic epigenetic upregulate Foxp3 gene [193]. Mechanically, using single T-cell analysis of RNA-seq, TCR tracking STARTRAC identified new regulatory mechanism and therapeutic strategy to reveal connections among different T-cell subsets (IFNG+ TH 1-like which IFN-γ-regulating transcription factors), CXCL13+ BHLHE40+ TH 1-like cells enriched in microsatellite instable MSI tumors [347]. On the other hand, MHCII+ Lgr5+ ISCs of TH cells modulate ISCs renewal and differentiation in opposing ways [348]. Besides, conversion to Lgr5+ ISCs and ISC dedifferentiation restore function upon external stimuli and intestinal homeostasis [349,350].
The microenvironment of tumor contains various cell types that include lymphocyte, blood vessel, fibroblastic cell, inflammatory cell and bone marrow cell [351]. Cancer association with chronic inflammation involved in T-cell [139], neoantigen-specific T cell activated in cancer immunotherapy indicating highly protective [97], but suppressive element in non-transformed epithelial cells (stromal cells) in regenerating tissues of the tumor microenvironment TME mediated restriction by excluding T cells indicating effective immunotherapy [351]. Neoantigen recognized may provide effective clinical immunotherapy [97], on which TME term immune suppressive element. Effectiveness of immune-suppressive TME further verify with the increase frequency of cancer-specific T cells [97,352], whereas loss of function of MDR1 were identified in a subset of ileal CD indicating that interaction between conjugated bile acids and Teff cells can regulate intestinal immune homeostasis [353]. Of note, mucosal healing of cholestyramine CME induced in primary sclerosing cholangitis-associated with IBD [354].
B–cells
B-cells subset Breg identified plays suppressive role on the immune system. As ATM response suppression, IL-21 induces granzyme B (GrB+) expression by B-cells in human Breg triggered by BCR and TLR signaling pathways [355,356]. Interestingly, TRAP (tumor cellreleased autophagosome) activates TLR2-MyD88-NF-KB signaling pathways in B cell induced by Breg cells differentiation into IL-10- produce B cell by HMGB1 on intact TRAPs [301], suggesting TRAP function in the induction of immunosuppression associate with development of cancer. Noticeably, the intrahepatic abundance of B-cell at tumor margin, Breg circulation (migrate from blood into tumor) promote tumor progression in HCC which significantly correlate with tumor invasiveness and stage. Breg induce cell proliferation and directly interaction with liver tumor cells through CD40/CD154 signaling pathways [357].
Here, PD-1high B cells identified as immunosuppressive cells induced in the microenvironment of HCC, whereas Breg cells interaction between PD-1/PD-LI induced dysfunction of T cells in different pathways either by PD-1 trigger on T- cell or regulatory B-cell produce IL-10 thereby induce effector T- cell dysfunction [358]. The immune blocker PD-1-PD-L1 interaction shows type 1 IFN activation as target for immunotherapy with high beneficial therapeutic [311]. Indeed, PD-1-PD-L1 interactions as new mechanism induce immune dysfunction during tumor progression. However, PD-1hi (Protumorigenic) B-cells trigger T-cell dysfunction and thereby cause progression of tumor via IL-10 dependent pathway [359]. B-cells FcγRIIlow/- activation prohibit T-cell immunity via IL-10 signals, result establish possible pathways which induce T-cell suppress and create possible avenue for tumor progression [299], whereas, activation of T-cell caused severe impairment to the intestine and intestinal barrier disruption and apoptosis [300]. Thus, provided mechanism by which immune responses regulate gut microbiome and epithelium [346]. Recently, GUDCA (as intestinal FXR antagonist) improve metabolic dysfunction of Bacteroides fragilis-GUDCA-intestinal FXR axis via an AMPK-independent mechanism, thus increased GUDCA level by decreasing B.fragilis inhibiting intestinal FXR signaling by hepatic CYP7A1 activity in the gut [159]. Cancer epigenetic silencing (DNA methylation and histone modification) is an important tumorigenic mechanism that represses tumor production of Th1-type chemokine CXCL9 and CXCL10 and enhance clinical efficacy of cancer therapy [360]. The protumorigenic role of CXCR3+ as a link between proinflammatory response and immunity cancer microenvironment confirmed, proposed IL-17+ cells might be regulator for CXCR3+ B-cells trafficking to cancers [361]. Notwithstanding that, CXCR3+ B-cells promote recruitment and accumulation at the invading edge of HCC. B-lymphocytes plays role in cancer progression, of which TNFα-producing B-cells limit senescence-mediated fibrosis and favors HCC progression. In established model of Mdr2-/- mice, showed distinctive B- and T- cell role of hepatic fibrogenesis, which B-Mdr2-/- inhibited fibrosis and accumulate senescence inactivation of hepatic stellate cell Hsc [362,363]. Correspondingly, effect of immunosuppressive drug, mycophenolate mofetil MMF inhibited inosine monophosphate dehydrogenase irreversibly dependent to T and B-lymphocytes [364].
HLA cells
HeLa cells commonly been used in human cell line, which first identified since 1952 [365]. Study of identified using PCR-based assay to detect Hela that are highly specific for HeLa [366]. Various immune response genes (HLA type and polymorphisms) reported as cause of clearance in HCV [312]. The complex found in HeLa cell contained TRIM24 and TRIM 33, of note revealed in purification of TRIM24 revealed TRIM28 and TRIM 33 although are more closely related towards HPI and HDACs 1 and 11 proteins [20,21]. Recent study found genetic variant on 13 loci across HLA class 11 associated strongly with PBC [367]. Genome wide association study with help of their technology identified the risk loci as HLA confirmed being the strongest and others like IL12, IL12RB2, 17q12.21,SP1B, CTLA-4, STAT4, MMEL1, IRF5- TNPO3, immune pathway involve in T- cell differentiation, B- cell function, antigen presentation and myeloid cell differentiation in PBC [85]. Importantly, HLA associated been strongly implicated the adaptive immune system on chromosome 6 is that encode class 1 and 11 complexes, that linked HLA-type of cellular phenotype to PBC noting potential linking genetic data with clinical outcome and disease subtype . Study indicated interleukin -12 immunoregulatory signaling axis directly related to PBC pathophysiology, associated between PBC and genetic variant at HLA class 11 region, IL12, IL12A and 1L12RB2 loci [367-368]. The link between HLA allele and PBC has been reported [82-84], risk allele occur in gene associated with immune function that intersect in various immune pathways, that has been identified in GWAs study as B-cell function, T-cells differentiation, antigen presenting and myeloid cell differentiation [85]. Of note that three loci gene in TNF-α signaling pathway identified by GWA study: TNFRSFIA, DENND1B, TNFAIP2 [82,86]. Study identifies HLA-C1 ligand gene KIR2DL3 inhibitory NK cells receptors to influence HCV in liver disease [369], which is necessary for determining antiviral immunity.
Discussion
The gut microbiome has intimate interaction with host immune system influenced by innate and adaptive immune function [221], which include neutrophil, DC, NK cell. The development of T-cell subtypes associated to a particular microbiome [222], of which adaptive immune response involve in B and T-cell activation. LPS identified as a triggering factor situated at blood stream from gut at the early development of metabolic endotoxemia disease of which insulin resistance associate with low-grade inflammation [223]. Disruption to the gut wall (tight junction) resulted in alteration. The gut wall becomes leaky, LPS goes into the blood stream. Excessively decrease the inflammatory tone and trigger glucose metabolism [223]. Given that gut microbiome function and bacterial composition change associated with dysbiosis results in an increase oxidative stress thereby, motivate a chronic inflammatory response [224]. Besides, in an ability to exacerbate inflammation with a reduction produced short chain fatty acid, whereas deficiency of SCFA in the epithelium raise in inflammatory response [226]. The metabolism of microbiome can modify by the host genetic, environmental factor based on dietary consumption and most changes are reversible although microbiota abundance depends on diet history [206,227-229]. Shaping individual microbiome associated with complex polygenic traits that affect microbiota composition in three ways [228], and change in BAs. TGR5 mRNA expression detect at increase level in small intestine, liver, stomach and lungs [257,271]. The innate and adaptive immune cell activities associated to liver cancer development, given that liver exposed to P/MAMP through PRR (TLR, NLR), which were confirmed [17]. The loss of negatively regulation of TLR signaling associated with autoimmune and inflammatory pathogenesis thus plays role in host defense and diseases [281,282]. Furthermore, T-cells induced mucosal damage by perforin via combined effect of different pathways of cytotoxicity like IFN-γ, TNF-α, perforin and Fas/Fasl Fas ligand (In regulation of immune system and cancer progression) [300].
Trim family protein directly interacts with cellular proteins as a combined effect or single thereby modulate signaling pathways that triggers engagement of PRR. Expression of pro-inflammatory responses (cytokine) affects IFNs (type I and II) by downstream regulation, also promote adaptive immune responses [19]. The role of Trim family protein established in innate immunity, differentiation, migration, proliferation, apoptosis, tumor development and progression [237,238]. More especially, its involvement in biological processes such as growth factors, immune cells [237,239-241,328,329]. Trim24 involved in p53 regulation whereas Trim28 in p53 inactivation, thus highly expresses in cancer [238]. Trim33 prevent the onset development of human myelomonocytic leukemia [53]. Studies reported of relationship between Trim family proteins and autophagy [313-315].
To attend accuracy in liver disease prognosis remain challenging [122]. Genome wide investigation shows that demethylation occurs simultaneously with tumor progression and activation of transposable element via genome wide hypomethylation in HCC [133]. It has established gut microbiome and immune cell play roles in various liver disease developments. Researches have been going on using different technologies in order to unravel the link and prognosis of this disease in human. Of note, gut microbiome acted through BA metabolism to regulate liver cancer via NKT cells [162]. Despite role of BA regulation of liver cancer and modulating FXR signaling on beneficial effect on the intestinal barrier and immune function [286]. The gut microbiome use BA shape immunity of liver cancer. More so they do by altering the gut microflora thereby bring about changes between anti-tumor immunity and hepatic immune cells against tumors in liver cancer [162]. Cholestasis induce liver injury cause inflammation by alteration of BAs profiles [104]. Variant rs72613567: TA HSD17B13 associate with reduce risk of liver disease and NASH progression [94]. Alteration of Trim28-dependent transcriptional dynamics due to obesity and aging precipitate, thus leads to metabolic infection [49]. Trim28 induced by interferon to resistance of pathogen. Better understanding of PRRs (innate and adaptive immune response) needed in therapeutic and drugs development for cancer and inflammatory diseases. Since PRR signaling pathway plays role in tolerance, pathogen elimination associated in inflammatory diseases and cancer. Study suggested that OCA improved in NASH histological features indicating long-term benefit and safety of obeticholic acid OCA need further clarification [8], long-term safety, efficacy (10 mg OCA) as novel therapy in PBC patient were supported [72], whereas 25-50 mg OCA for 6 weeks was tolerated [120]. Acid dose-dependent increased incidence of pruritus with obeticholic acid, concomitant administration of obeticholic acid OCA with ursodiol or alone shows improved ALP, bilirubin level in PBC patients except pruritus no serious adverse events seen [80]. Suggesting that 10 mg of OCA to be effective dose, long-term safety, efficacy of OCA in PBC patient support 5-fold range of OCA, non-clear differences in biochemical end-point observed indicated doses to be high for trial [72]. In as much as decrease marker of liver inflammation, fibrosis with T2D and NAFLD patients, an increase insulin sensitivity observed with 25–50 mg OCA administration for 6 weeks tolerated [120]. However, UDCA being only drug approve for PBC treatment, suggested use of noninvasive marker to monitor the disease progression [258]. The time for disease development according to fibrosis stage can be included in counseling and decision of patient’s health [122]. According to one study, growth hormone may be novel target for development of liver fibrosis therapeutic [81]. More so, JTE-013 showed therapeutic target for cholestatic liver diseases [264]. The growth hormone resistance may have play role or cause onset, development of Inflammatory Cholestasis induce liver fibrosis in mice model [81]. In all these, what is the way out of this deadly disease without adverse events and relapses, whereas the role diet and chemical play in disease management need to put under consideration.
REFERENCES
- Global, regional and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet (London, England). 2015;386(9995):743-800.
- Global, regional and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet (London, England). 2015;385(9963):117-71.
- Fitzmaurice C, Dicker D, Pain A, Hamavid H, Moradi-Lakeh M, MacIntyre MF, et al. The Global Burden of Cancer 2013. JAMA oncology. 2015;1(4):505-27.
- Sanyal AJ. AGA technical review on nonalcoholic fatty liver disease. Gastroenterology. 2002;123(5):1705-25.
- Chazouilleres O, Wendum D, Serfaty L, Montembault S, Rosmorduc O, Poupon R. Primary biliary cirrhosis-autoimmune hepatitis overlap syndrome: clinical features and response to therapy. Hepatology (Baltimore, Md). 1998;28(2):296-301.
- Hardy J, Singleton A. Genomewide association studies and human disease. The New England journal of medicine. 2009;360(17):1759-68.
- Dawkins J, Webster RM. The hepatocellular carcinoma market. Nature reviews Drug discovery. 2018.
- Neuschwander-Tetri BA, Loomba R, Sanyal AJ, Lavine JE, Van Natta ML, Abdelmalek MF, et al. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): a multicentre, randomised, placebo-controlled trial. Lancet (London, England). 2015;385(9972):956-65.
- Anstee QM, Daly AK, Day CP. Genetics of Alcoholic Liver Disease. Seminars in liver disease. 2015;35(4):361-74.
- Anstee QM, Day CP. The Genetics of Nonalcoholic Fatty Liver Disease: Spotlight on PNPLA3 and TM6SF2. Seminars in liver disease. 2015;35(3):270-90.
- Anstee QM, Seth D, Day CP. Genetic Factors That Affect Risk of Alcoholic and Nonalcoholic Fatty Liver Disease. Gastroenterology. 2016;150(8):1728-44.e7.
- Hirschhorn JN, Gajdos ZK. Genome-wide association studies: results from the first few years and potential implications for clinical medicine. Annual review of medicine. 2011;62:11-24.
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747-53.
- Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nature genetics. 2009;41(1):25-34.
- Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455(7210):232-6.
- Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460(7256):744-7.
- Kawai T, Akira S. Regulation of innate immune signalling pathways by the tripartite motif (TRIM) family proteins. EMBO molecular medicine. 2011;3(9):513-27.
- Carthagena L, Bergamaschi A, Luna JM, David A, Uchil PD, Margottin-Goguet F, et al. Human TRIM gene expression in response to interferons. PloS one. 2009;4(3):e4894.
- Ozato K, Shin DM, Chang TH, Morse HC, 3rd. TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 2008;8(11):849-60.
- Herquel B, Ouararhni K, Davidson I. The TIF1alpha-related TRIM cofactors couple chromatin modifications to transcriptional regulation, signaling and tumor suppression. Transcription. 2011;2(5):231-6.
- Herquel B, Ouararhni K, Khetchoumian K, Ignat M, Teletin M, Mark M, et al. Transcription cofactors TRIM24, TRIM28, and TRIM33 associate to form regulatory complexes that suppress murine hepatocellular carcinoma. Proc Natl Acad Sci U S A. 2011;108(20):8212-7.
- Boekhoudt GH, Frazier-Jessen MR, Feldman GM. Immune complexes suppress IFN-gamma signaling by activation of the FcgammaRI pathway. Journal of leukocyte biology. 2007;81(4):1086-92.
- Buch S, Stickel F, Trepo E, Way M, Herrmann A, Nischalke HD, et al. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nature genetics. 2015;47(12):1443-8.
- Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nature reviews Gastroenterology & hepatology. 2018;15(1):11-20.
- Krawczyk M, Rau M, Schattenberg JM, Bantel H, Pathil A, Demir M, et al. Combined effects of the PNPLA3 rs738409, TM6SF2 rs58542926, and MBOAT7 rs641738 variants on NAFLD severity: a multicenter biopsy-based study. Journal of lipid research. 2017;58(1):247-55.
- Oleksiewicz U, Gladych M, Raman AT, Heyn H, Mereu E, Chlebanowska P, et al. TRIM28 and Interacting KRAB-ZNFs Control Self-Renewal of Human Pluripotent Stem Cells through Epigenetic Repression of Pro-differentiation Genes. Stem cell reports. 2017;9(6):2065-80.
- Wolf D, Goff SP. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell. 2007;131(1):46-57.
- Wolf G, Yang P, Fuchtbauer AC, Fuchtbauer EM, Silva AM, Park C, et al. The KRAB zinc finger protein ZFP809 is required to initiate epigenetic silencing of endogenous retroviruses. Genes & development. 2015;29(5):538-54.
- Khetchoumian K, Teletin M, Tisserand J, Mark M, Herquel B, Ignat M, et al. Loss of Trim24 (Tif1alpha) gene function confers oncogenic activity to retinoic acid receptor alpha. Nature genetics. 2007;39(12):1500-6.
- Tsai WW, Wang Z, Yiu TT, Akdemir KC, Xia W, Winter S, et al. TRIM24 links a non-canonical histone signature to breast cancer. Nature. 2010;468(7326):927-32.
- Bojkowska K, Aloisio F, Cassano M, Kapopoulou A, Santoni de Sio F, Zangger N, et al. Liver-specific ablation of Kruppel-associated box-associated protein 1 in mice leads to male-predominant hepatosteatosis and development of liver adenoma. Hepatology (Baltimore, Md). 2012;56(4):1279-90.
- Friedman JR, Fredericks WJ, Jensen DE, Speicher DW, Huang XP, Neilson EG, et al. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes & development. 1996;10(16):2067-78.
- Rauwel B, Jang SM, Cassano M, Kapopoulou A, Barde I, Trono D. Release of human cytomegalovirus from latency by a KAP1/TRIM28 phosphorylation switch. eLife. 2015;4.
- White DE, Negorev D, Peng H, Ivanov AV, Maul GG, Rauscher FJ, 3rd. KAP1, a novel substrate for PIKK family members, colocalizes with numerous damage response factors at DNA lesions. Cancer Res. 2006;66(24):11594-9.
- Le Douarin B, You J, Nielsen AL, Chambon P, Losson R. TIF1alpha: a possible link between KRAB zinc finger proteins and nuclear receptors. The Journal of steroid biochemistry and molecular biology. 1998;65(1-6):43-50.
- Le Douarin B, Nielsen AL, Garnier JM, Ichinose H, Jeanmougin F, Losson R, et al. A possible involvement of TIF1 alpha and TIF1 beta in the epigenetic control of transcription by nuclear receptors. The EMBO journal. 1996;15(23):6701-15.
- Le Douarin B, vom Baur E, Zechel C, Heery D, Heine M, Vivat V, et al. Ligand-dependent interaction of nuclear receptors with potential transcriptional intermediary factors (mediators). Philosophical transactions of the Royal Society of London Series B, Biological sciences. 1996;351(1339):569-78.
- Khetchoumian K, Teletin M, Tisserand J, Herquel B, Ouararhni K, Losson R. Trim24 (Tif1 alpha): an essential 'brake' for retinoic acid-induced transcription to prevent liver cancer. Cell cycle (Georgetown, Tex). 2008;7(23):3647-52.
- Villanueva A, Newell P, Chiang DY, Friedman SL, Llovet JM. Genomics and signaling pathways in hepatocellular carcinoma. Seminars in liver disease. 2007;27(1):55-76.
- Laurent-Puig P, Legoix P, Bluteau O, Belghiti J, Franco D, Binot F, et al. Genetic alterations associated with hepatocellular carcinomas define distinct pathways of hepatocarcinogenesis. Gastroenterology. 2001;120(7):1763-73.
- Laurent-Puig P, Zucman-Rossi J. Genetics of hepatocellular tumors. Oncogene. 2006;25(27):3778-86.
- Cai RL, Meng W, Lu HY, Lin WY, Jiang F, Shen FM. Segregation analysis of hepatocellular carcinoma in a moderately high-incidence area of East China. World journal of gastroenterology. 2003;9(11):2428-32.
- Harada H, Nagai H, Ezura Y, Yokota T, Ohsawa I, Yamaguchi K, et al. Down-regulation of a novel gene, DRLM, in human liver malignancy from 4q22 that encodes a NAP-like protein. Gene. 2002;296(1-2):171-7.
- White D, Rafalska-Metcalf IU, Ivanov AV, Corsinotti A, Peng H, Lee SC, et al. The ATM substrate KAP1 controls DNA repair in heterochromatin: regulation by HP1 proteins and serine 473/824 phosphorylation. Molecular cancer research : MCR. 2012;10(3):401-14.
- Yang J, Yu Y, Hamrick HE, Duerksen-Hughes PJ. ATM, ATR and DNA-PK: initiators of the cellular genotoxic stress responses. Carcinogenesis. 2003;24(10):1571-80.
- Lechner MS, Begg GE, Speicher DW, Rauscher FJ, 3rd. Molecular determinants for targeting heterochromatin protein 1-mediated gene silencing: direct chromoshadow domain-KAP-1 corepressor interaction is essential. Molecular and cellular biology. 2000;20(17):6449-65.
- Chang CW, Chou HY, Lin YS, Huang KH, Chang CJ, Hsu TC, et al. Phosphorylation at Ser473 regulates heterochromatin protein 1 binding and corepressor function of TIF1beta/KAP1. BMC molecular biology. 2008;9:61.
- Singh K, Cassano M, Planet E, Sebastian S, Jang SM, Sohi G, et al. A KAP1 phosphorylation switch controls MyoD function during skeletal muscle differentiation. Genes & development. 2015;29(5):513-25.
- Cassano M, Offner S, Planet E, Piersigilli A, Jang SM, Henry H, et al. Polyphenic trait promotes liver cancer in a model of epigenetic instability in mice. Hepatology (Baltimore, Md). 2017;66(1):235-51.
- Chassaing B, Etienne-Mesmin L, Gewirtz AT. Microbiota-liver axis in hepatic disease. Hepatology (Baltimore, Md). 2014;59(1):328-39.
- Wang E, Kawaoka S, Roe JS, Shi J, Hohmann AF, Xu Y, et al. The transcriptional cofactor TRIM33 prevents apoptosis in B lymphoblastic leukemia by deactivating a single enhancer. eLife. 2015;4:e06377.
- Gatt ME, Takada K, Mani M, Lerner M, Pick M, Hideshima T, et al. TRIM13 (RFP2) downregulation decreases tumour cell growth in multiple myeloma through inhibition of NF Kappa B pathway and proteasome activity. Br J Haematol. 2013;162(2):210-20.
- Jackson CC, Medeiros LJ, Miranda RN. 8p11 myeloproliferative syndrome: a review. Human pathology. 2010;41(4):461-76.
- Quintas-Cardama A, Zhang N, Qiu YH, Post S, Creighton CJ, Cortes J, et al. Loss of TRIM62 expression is an independent adverse prognostic factor in acute myeloid leukemia. Clinical lymphoma, myeloma & leukemia. 2015;15(2):115-27.e15.
- Aucagne R, Droin N, Paggetti J, Lagrange B, Largeot A, Hammann A, et al. Transcription intermediary factor 1gamma is a tumor suppressor in mouse and human chronic myelomonocytic leukemia. J Clin Invest. 2011;121(6):2361-70.
- Crawford LJ, Johnston CK, Irvine AE. TRIM proteins in blood cancers. Journal of cell communication and signaling. 2018;12(1):21-9.
- Kurahashi S, Hayakawa F, Miyata Y, Yasuda T, Minami Y, Tsuzuki S, et al. PAX5-PML acts as a dual dominant-negative form of both PAX5 and PML. Oncogene. 2011;30(15):1822-30.
- Bai X, Kim J, Yang Z, Jurynec MJ, Akie TE, Lee J, et al. TIF1gamma controls erythroid cell fate by regulating transcription elongation. Cell. 2010;142(1):133-43.
- Bai X, Trowbridge JJ, Riley E, Lee JA, DiBiase A, Kaartinen VM, et al. TiF1-gamma plays an essential role in murine hematopoiesis and regulates transcriptional elongation of erythroid genes. Developmental biology. 2013;373(2):422-30.
- Demy DL, Tauzin M, Lancino M, Le Cabec V, Redd M, Murayama E, et al. Trim33 is essential for macrophage and neutrophil mobilization to developmental or inflammatory cues. Journal of cell science. 2017;130(17):2797-807.
- Agricola E, Randall RA, Gaarenstroom T, Dupont S, Hill CS. Recruitment of TIF1gamma to chromatin via its PHD finger-bromodomain activates its ubiquitin ligase and transcriptional repressor activities. Molecular cell. 2011;43(1):85-96.
- Li F, Hu L, Xu Y, Li Z, Yi S, Gu Z, et al. Identification of characteristic and prognostic values of chromosome 1p abnormality by multi-gene fluorescence in situ hybridization in multiple myeloma. Leukemia. 2016;30(5):1197-201.
- Walker BA, Leone PE, Chiecchio L, Dickens NJ, Jenner MW, Boyd KD. A compendium of myeloma-associated chromosomal copy number abnormalities and their prognostic value. Blood. 2010;116:e56-e65.
- Li F, Xu Y, Deng P, Yang Y, Sui W, Jin F. Heterogeneous chromosome 12p deletion is an independent adverse prognostic factor and resistant to bortezomib-based therapy in multiple myeloma. Oncotarget. 2015;6:9434-44.
- Hirschfield GM, Gershwin ME. The immunobiology and pathophysiology of primary biliary cirrhosis. Annu Rev Pathol. 2013;8:303-30.
- Poupon RE, Chretien Y, Chazouilleres O, Poupon R, Chwalow J. Quality of life in patients with primary biliary cirrhosis. Hepatology (Baltimore, Md). 2004;40(2):489-94.
- Talwalkar JA, Souto E, Jorgensen RA, Lindor KD. Natural history of pruritus in primary biliary cirrhosis. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2003;1(4):297-302.
- Colina F, Pinedo F, Solis JA, Moreno D, Nevado M. Nodular regenerative hyperplasia of the liver in early histological stages of primary biliary cirrhosis. Gastroenterology. 1992;102(4 Pt 1):1319-24.
- Longo M, Crosignani A, Battezzati PM, Squarcia Giussani C, Invernizzi P, Zuin M, et al. Hyperlipidaemic state and cardiovascular risk in primary biliary cirrhosis. Gut. 2002;51(2):265-9.
- Menon KV, Angulo P, Weston S, Dickson ER, Lindor KD. Bone disease in primary biliary cirrhosis: independent indicators and rate of progression. Journal of hepatology. 2001;35(3):316-23.
- Munoz SJ, Heubi JE, Balistreri WF, Maddrey WC. Vitamin E deficiency in primary biliary cirrhosis: gastrointestinal malabsorption, frequency and relationship to other lipid-soluble vitamins. Hepatology (Baltimore, Md). 1989;9(4):525-31.
- Hirschfield GM, Mason A, Luketic V, Lindor K, Gordon SC, Mayo M, et al. Efficacy of obeticholic acid in patients with primary biliary cirrhosis and inadequate response to ursodeoxycholic acid. Gastroenterology. 2015;148(4):751-61.e8.
- Lindor KD, Gershwin ME, Poupon R, Kaplan M, Bergasa NV, Heathcote EJ. Primary biliary cirrhosis. Hepatology (Baltimore, Md). 2009;50(1):291-308.
- Dyson JK, Hirschfield GM, Adams DH, Beuers U, Mann DA, Lindor KD, et al. Novel therapeutic targets in primary biliary cirrhosis. Nature reviews Gastroenterology & hepatology. 2015;12(3):147-58.
- Gershwin ME, Selmi C, Worman HJ, Gold EB, Watnik M, Utts J, et al. Risk factors and comorbidities in primary biliary cirrhosis: a controlled interview-based study of 1032 patients. Hepatology (Baltimore, Md). 2005;42(5):1194-202.
- Hamlyn AN, Macklon AF, James O. Primary biliary cirrhosis: geographical clustering and symptomatic onset seasonality. Gut. 1983;24(10):940-5.
- Nakamura M. Clinical significance of autoantibodies in primary biliary cirrhosis. Seminars in liver disease. 2014;34(3):334-40.
- Frazer IH, Mackay IR, Jordan TW, Whittingham S, Marzuki S. Reactivity of anti-mitochondrial autoantibodies in primary biliary cirrhosis: definition of two novel mitochondrial polypeptide autoantigens. The Journal of Immunology. 1985;135(3):1739-45.
- Gossard AA, Lindor KD. Development of autoimmune hepatitis in primary biliary cirrhosis. Liver international : official journal of the International Association for the Study of the Liver. 2007;27(8):1086-90.
- Nevens F, Andreone P, Mazzella G, Strasser SI, Bowlus C, Invernizzi P, et al. A Placebo-Controlled Trial of Obeticholic Acid in Primary Biliary Cholangitis. The New England journal of medicine. 2016;375(7):631-43.
- Stiedl P, McMahon R, Blaas L, Stanek V, Svinka J, Grabner B, et al. Growth hormone resistance exacerbates cholestasis-induced murine liver fibrosis. Hepatology (Baltimore, Md). 2015;61(2):613-26.
- Nakamura M. [Analysis of disease-pathway by identifying susceptible genes to primary biliary cirrhosis]. Nihon Rinsho Men'eki Gakkai kaishi = Japanese journal of clinical immunology. 2012;35(6):503-10.
- Invernizzi P. Human leukocyte antigen in primary biliary cirrhosis: an old story now reviving. Hepatology (Baltimore, Md). 2011;54(2):714-23.
- Invernizzi P, Battezzati PM, Crosignani A, Perego F, Poli F, Morabito A, et al. Peculiar HLA polymorphisms in Italian patients with primary biliary cirrhosis. Journal of hepatology. 2003;38(4):401-6.
- Carbone M, Lleo A, Sandford RN, Invernizzi P. Implications of genome-wide association studies in novel therapeutics in primary biliary cirrhosis. European journal of immunology. 2014;44(4):945-54.
- Mells GF, Floyd JA, Morley KI, Cordell HJ, Franklin CS, Shin SY, et al. Genome-wide association study identifies 12 new susceptibility loci for primary biliary cirrhosis. Nature genetics. 2011;43(4):329-32.
- Abdelmalek MF, Liu C, Shuster J, Nelson DR, Asal NR. Familial aggregation of insulin resistance in first-degree relatives of patients with nonalcoholic fatty liver disease. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2006;4(9):1162-9.
- Pagliaro L, Pasta L, D'Amico G, Madonia S, Pietrosi G. Familial clustering of (mostly) HCV-related cirrhosis. A case-control study. Journal of hepatology. 2002;37(6):762-6.
- Yu MW, Chang HC, Liaw YF, Lin SM, Lee SD, Liu CJ, et al. Familial risk of hepatocellular carcinoma among chronic hepatitis B carriers and their relatives. Journal of the National Cancer Institute. 2000;92(14):1159-64.
- Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nature genetics. 2008;40(12):1461-5.
- Kozlitina J, Smagris E, Stender S, Nordestgaard BG, Zhou HH, Tybjaerg-Hansen A, et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nature genetics. 2014;46(4):352-6.
- Kitamoto T, Kitamoto A, Yoneda M, Hyogo H, Ochi H, Nakamura T, et al. Genome-wide scan revealed that polymorphisms in the PNPLA3, SAMM50, and PARVB genes are associated with development and progression of nonalcoholic fatty liver disease in Japan. Human genetics. 2013;132(7):783-92.
- Feitosa MF, Wojczynski MK, North KE, Zhang Q, Province MA, Carr JJ, et al. The ERLIN1-CHUK-CWF19L1 gene cluster influences liver fat deposition and hepatic inflammation in the NHLBI Family Heart Study. Atherosclerosis. 2013;228(1):175-80.
- Abul-Husn NS, Cheng X, Li AH, Xin Y, Schurmann C, Stevis P, et al. A Protein-Truncating HSD17B13 Variant and Protection from Chronic Liver Disease. The New England journal of medicine. 2018;378(12):1096-106.
- Bosch A, Dumortier J, Maucort-Boulch D, Scoazec JY, Wendum D, Conti F, et al. Preventive administration of UDCA after liver transplantation for primary biliary cirrhosis is associated with a lower risk of disease recurrence. Journal of hepatology. 2015;63(6):1449-58.
- Singal AG, El-Serag HB. Hepatocellular Carcinoma From Epidemiology to Prevention: Translating Knowledge into Practice. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2015;13(12):2140-51.
- Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science (New York, NY). 2015;348(6230):69-74.
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646-74.
- <CDC. Viral Hepatitis Surveillance - United State, 2016 www.cdc.govhepatitis B virus.pdf>.
- Klevens RM, Liu S, Roberts H, Jiles RB, Holmberg SD. Estimating acute viral hepatitis infections from nationally reported cases. American journal of public health. 2014;104(3):482-7.
- <CDC. Viral Hepatitis Surveillance - United State, 2016 www.cdc.govhepatitis C virus.pdf>.
- Rosenberg ES, Rosenthal EM, Hall EW, Barker L, Hofmeister MG, Sullivan PS, et al. Prevalence of Hepatitis C Virus Infection in US States and the District of Columbia, 2013 to 2016. JAMA network open. 2018;1(8):e186371.
- Hofmeister MG, Rosenthal EM, Barker LK, Rosenberg ES, Barranco MA, Hall EW, et al. Estimating Prevalence of Hepatitis C Virus Infection in the United States, 2013-2016. Hepatology (Baltimore, Md). 2019;69(3):1020-31.
- Zhang Y, Shi Y, Wu R, Wang X, Gao X, Niu J. Primary biliary cholangitis is more severe in previous hepatitis B virus infection patients. European journal of gastroenterology & hepatology. 2018;30(6):682-6.
- Mehta SH, Cox A, Hoover DR, Wang XH, Mao Q, Ray S, et al. Protection against persistence of hepatitis C. Lancet (London, England). 2002;359(9316):1478-83.
- Grebely J, Prins M, Hellard M, Cox AL, Osburn WO, Lauer G, et al. Hepatitis C virus clearance, reinfection, and persistence, with insights from studies of injecting drug users: towards a vaccine. The Lancet Infectious diseases. 2012;12(5):408-14.
- Wong VW, Adams LA, de Ledinghen V, Wong GL, Sookoian S. Noninvasive biomarkers in NAFLD and NASH - current progress and future promise. Nature reviews Gastroenterology & hepatology. 2018;15(8):461-78.
- Cohen JC, Horton JD, Hobbs HH. Human fatty liver disease: old questions and new insights. Science (New York, NY). 2011;332(6037):1519-23.
- Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology (Baltimore, Md). 2016;64(1):73-84.
- Williams CD, Stengel J, Asike MI, Torres DM, Shaw J, Contreras M, et al. Prevalence of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis among a largely middle-aged population utilizing ultrasound and liver biopsy: a prospective study. Gastroenterology. 2011;140(1):124-31.
- Browning JD, Szczepaniak LS, Dobbins R, Nuremberg P, Horton JD, Cohen JC, et al. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology (Baltimore, Md). 2004;40(6):1387-95.
- Younossi ZM, Stepanova M, Afendy M, Fang Y, Younossi Y, Mir H, et al. Changes in the prevalence of the most common causes of chronic liver diseases in the United States from 1988 to 2008. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2011;9(6):524-30.e1; quiz e60.
- Bazick J, Donithan M, Neuschwander-Tetri BA, Kleiner D, Brunt EM, Wilson L, et al. Clinical Model for NASH and Advanced Fibrosis in Adult Patients With Diabetes and NAFLD: Guidelines for Referral in NAFLD. Diabetes care. 2015;38(7):1347-55.
- Cusi K, Sanyal AJ, Zhang S, Hartman ML, Bue-Valleskey JM, Hoogwerf BJ, et al. Non-alcoholic fatty liver disease (NAFLD) prevalence and its metabolic associations in patients with type 1 diabetes and type 2 diabetes. Diabetes, obesity & metabolism. 2017;19(11):1630-4.
- Liu X, Huang Y, Yang D, Li X, Liang J, Lin L, et al. Overexpression of TRIM24 is associated with the onset and progress of human hepatocellular carcinoma. PloS one. 2014;9(1):e85462.
- Adams LA, Lymp JF, St Sauver J, Sanderson SO, Lindor KD, Feldstein A, et al. The natural history of nonalcoholic fatty liver disease: a population-based cohort study. Gastroenterology. 2005;129(1):113-21.
- Ascha MS, Hanouneh IA, Lopez R, Tamimi TA, Feldstein AF, Zein NN. The incidence and risk factors of hepatocellular carcinoma in patients with nonalcoholic steatohepatitis. Hepatology (Baltimore, Md). 2010;51(6):1972-8.
- Valenti L, Al-Serri A, Daly AK, Galmozzi E, Rametta R, Dongiovanni P, et al. Homozygosity for the patatin-like phospholipase-3/adiponutrin I148M polymorphism influences liver fibrosis in patients with nonalcoholic fatty liver disease. Hepatology (Baltimore, Md). 2010;51(4):1209-17.
- Liu YL, Reeves HL, Burt AD, Tiniakos D, McPherson S, Leathart JB, et al. TM6SF2 rs58542926 influences hepatic fibrosis progression in patients with non-alcoholic fatty liver disease. Nature communications. 2014;5:4309.
- Mudaliar S, Henry RR, Sanyal AJ, Morrow L, Marschall HU, Kipnes M, et al. Efficacy and safety of the farnesoid X receptor agonist obeticholic acid in patients with type 2 diabetes and nonalcoholic fatty liver disease. Gastroenterology. 2013;145(3):574-82.e1.
- McPherson S, Hardy T, Henderson E, Burt AD, Day CP, Anstee QM. Evidence of NAFLD progression from steatosis to fibrosing-steatohepatitis using paired biopsies: implications for prognosis and clinical management. Journal of hepatology. 2015;62(5):1148-55.
- Hagstrom H, Nasr P, Ekstedt M, Hammar U, Stal P, Hultcrantz R, et al. Fibrosis stage but not NASH predicts mortality and time to development of severe liver disease in biopsy-proven NAFLD. Journal of hepatology. 2017;67(6):1265-73.
- Sorensen HT, Friis S, Olsen JH, Thulstrup AM, Mellemkjaer L, Linet M, et al. Risk of liver and other types of cancer in patients with cirrhosis: a nationwide cohort study in Denmark. Hepatology (Baltimore, Md). 1998;28(4):921-5.
- El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132(7):2557-76.
- Rudolph KL, Chang S, Millard M, Schreiber-Agus N, DePinho RA. Inhibition of experimental liver cirrhosis in mice by telomerase gene delivery. Science (New York, NY). 2000;287(5456):1253-8.
- Suzuki A, Lymp J, Donlinger J, Mendes F, Angulo P, Lindor K. Clinical predictors for hepatocellular carcinoma in patients with primary biliary cirrhosis. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2007;5(2):259-64.
- Trivedi PJ, Lammers WJ, van Buuren HR, Pares A, Floreani A, Janssen HL, et al. Stratification of hepatocellular carcinoma risk in primary biliary cirrhosis: a multicentre international study. Gut. 2016;65(2):321-9.
- Silveira MG, Suzuki A, Lindor KD. Surveillance for hepatocellular carcinoma in patients with primary biliary cirrhosis. Hepatology (Baltimore, Md). 2008;48(4):1149-56.
- Imam MH, Silveira MG, Sinakos E, Gossard AA, Jorgensen R, Keach J, et al. Long-term outcomes of patients with primary biliary cirrhosis and hepatocellular carcinoma. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2012;10(2):182-5.
- Takayama T, Makuuchi M, Hirohashi S, Sakamoto M, Okazaki N, Takayasu K, et al. Malignant transformation of adenomatous hyperplasia to hepatocellular carcinoma. Lancet (London, England). 1990;336(8724):1150-3.
- Gama-Sosa MA, Slagel VA, Trewyn RW, Oxenhandler R, Kuo KC, Gehrke CW, et al. The 5-methylcytosine content of DNA from human tumors. Nucleic acids research. 1983;11(19):6883-94.
- Bedford MT, van Helden PD. Hypomethylation of DNA in Pathological Conditions of the Human Prostate. Cancer Research. 1987;47(20):5274-6.
- Lin CH, Hsieh SY, Sheen IS, Lee WC, Chen TC, Shyu WC, et al. Genome-wide hypomethylation in hepatocellular carcinogenesis. Cancer Res. 2001;61(10):4238-43.
- Cao S, Zhu X, Zhang C, Qian H, Schuttler HB, Gong J, et al. Competition between DNA Methylation, Nucleotide Synthesis, and Antioxidation in Cancer versus Normal Tissues. Cancer Res. 2017;77(15):4185-95.
- Yang X, Gao L, Zhang S. Comparative pan-cancer DNA methylation analysis reveals cancer common and specific patterns. Briefings in bioinformatics. 2017;18(5):761-73.
- Nijhawan PK, Therneau TM, Dickson ER, Boynton J, Lindor KD. Incidence of cancer in primary biliary cirrhosis: the Mayo experience. Hepatology (Baltimore, Md). 1999;29(5):1396-8.
- Howel D, Metcalf JV, Gray J, Newman WL, Jones DE, James OF. Cancer risk in primary biliary cirrhosis: a study in northern England. Gut. 1999;45(5):756-60.
- Floreani A, Baragiotta A, Baldo V, Menegon T, Farinati F, Naccarato R. Hepatic and extrahepatic malignancies in primary biliary cirrhosis. Hepatology (Baltimore, Md). 1999;29(5):1425-8.
- Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883-99.
- Jones VS, Huang RY, Chen LP, Chen ZS, Fu L, Huang RP. Cytokines in cancer drug resistance: Cues to new therapeutic strategies. Biochimica et biophysica acta. 2016;1865(2):255-65.
- Tagliamonte M, Petrizzo A, Tornesello ML, Ciliberto G, Buonaguro FM, Buonaguro L. Combinatorial immunotherapy strategies for hepatocellular carcinoma. Current opinion in immunology. 2016;39:103-13.
- Zitvogel L, Pitt JM, Daillere R, Smyth MJ, Kroemer G. Mouse models in oncoimmunology. Nature reviews Cancer. 2016;16(12):759-73.
- Sprinzl MF, Galle PR. Current progress in immunotherapy of hepatocellular carcinoma. Journal of hepatology. 2017;66(3):482-4.
- Zhao Y, Shuen TWH, Toh TB, Chan XY, Liu M, Tan SY, et al. Development of a new patient-derived xenograft humanised mouse model to study human-specific tumour microenvironment and immunotherapy. Gut. 2018;67(10):1845-54.
- Lu X, Ye K, Zou K, Chen J. Identification of copy number variation-driven genes for liver cancer via bioinformatics analysis. Oncology reports. 2014;32(5):1845-52.
- Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451-81.
- Vissers LE, de Vries BB, Veltman JA. Genomic microarrays in mental retardation: from copy number variation to gene, from research to diagnosis. Journal of medical genetics. 2010;47(5):289-97.
- Inagawa S, Itabashi M, Adachi S, Kawamoto T, Hori M, Shimazaki J, et al. Expression and prognostic roles of beta-catenin in hepatocellular carcinoma: correlation with tumor progression and postoperative survival. Clin Cancer Res. 2002;8(2):450-6.
- Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441(7092):431-6.
- Karin M. NF-kappaB as a critical link between inflammation and cancer. Cold Spring Harbor perspectives in biology. 2009;1(5):a000141.
- DiDonato JA, Mercurio F, Karin M. NF-kappaB and the link between inflammation and cancer. Immunological reviews. 2012;246(1):379-400.
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124(4):783-801.
- Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1(2):135-45.
- Karin M, Cao Y, Greten FR, Li ZW. NF-kappaB in cancer: from innocent bystander to major culprit. Nature reviews Cancer. 2002;2(4):301-10.
- Mauad TH, van Nieuwkerk CM, Dingemans KP, Smit JJ, Schinkel AH, Notenboom RG, et al. Mice with homozygous disruption of the mdr2 P-glycoprotein gene. A novel animal model for studies of nonsuppurative inflammatory cholangitis and hepatocarcinogenesis. The American journal of pathology. 1994;145(5):1237-45.
- Maeda S, Kamata H, Luo JL, Leffert H, Karin M. IKKbeta couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell. 2005;121(7):977-90.
- Sommer F, Backhed F. The gut microbiota--masters of host development and physiology. Nature reviews Microbiology. 2013;11(4):227-38.
- Liu R, Zhao R, Zhou X, Liang X, Campbell DJ, Zhang X, et al. Conjugated bile acids promote cholangiocarcinoma cell invasive growth through activation of sphingosine 1-phosphate receptor 2. Hepatology (Baltimore, Md). 2014;60(3):908-18.
- Sun L, Xie C, Wang G, Wu Y, Wu Q, Wang X, et al. Gut microbiota and intestinal FXR mediate the clinical benefits of metformin. Nat Med. 2018;24(12):1919-29.
- Pathak P, Xie C, Nichols RG, Ferrell JM, Boehme S, Krausz KW, et al. Intestine farnesoid X receptor agonist and the gut microbiota activate G-protein bile acid receptor-1 signaling to improve metabolism. Hepatology (Baltimore, Md). 2018;68(4):1574-88.
- Hu X, Bonde Y, Eggertsen G, Rudling M. Muricholic bile acids are potent regulators of bile acid synthesis via a positive feedback mechanism. Journal of internal medicine. 2014;275(1):27-38.
- Ma C, Han M, Heinrich B, Fu Q, Zhang Q, Sandhu M, et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science (New York, NY). 2018;360(6391).
- Cadigan KM, Waterman ML. TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harbor perspectives in biology. 2012;4(11).
- Gao C, Xiao G, Hu J. Regulation of Wnt/beta-catenin signaling by posttranslational modifications. Cell & bioscience. 2014;4(1):13.
- Abrink M, Ortiz JA, Mark C, Sanchez C, Looman C, Hellman L, et al. Conserved interaction between distinct Kruppel-associated box domains and the transcriptional intermediary factor 1 beta. Proc Natl Acad Sci U S A. 2001;98(4):1422-6.
- vom Baur E, Zechel C, Heery D, Heine MJ, Garnier JM, Vivat V, et al. Differential ligand-dependent interactions between the AF-2 activating domain of nuclear receptors and the putative transcriptional intermediary factors mSUG1 and TIF1. The EMBO journal. 1996;15(1):110-24.
- Allton K, Jain AK, Herz HM, Tsai WW, Jung SY, Qin J, et al. Trim24 targets endogenous p53 for degradation. Proc Natl Acad Sci U S A. 2009;106(28):11612-6.
- Dupont S, Mamidi A, Cordenonsi M, Montagner M, Zacchigna L, Adorno M, et al. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell. 2009;136(1):123-35.
- He W, Dorn DC, Erdjument-Bromage H, Tempst P, Moore MA, Massague J. Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGFbeta pathway. Cell. 2006;125(5):929-41.
- Sherr CJ. Principles of tumor suppression. Cell. 2004;116(2):235-46.
- Caja L, Sancho P, Bertran E, Fabregat I. Dissecting the effect of targeting the epidermal growth factor receptor on TGF-beta-induced-apoptosis in human hepatocellular carcinoma cells. Journal of hepatology. 2011;55(2):351-8.
- Zhu H, Wu K, Yan W, Hu L, Yuan J, Dong Y, et al. Epigenetic silencing of DACH1 induces loss of transforming growth factor-beta1 antiproliferative response in human hepatocellular carcinoma. Hepatology (Baltimore, Md). 2013;58(6):2012-22.
- Caja L, Bertran E, Campbell J, Fausto N, Fabregat I. The transforming growth factor-beta (TGF-beta) mediates acquisition of a mesenchymal stem cell-like phenotype in human liver cells. Journal of cellular physiology. 2011;226(5):1214-23.
- Caja L, Sancho P, Bertran E, Ortiz C, Campbell JS, Fausto N, et al. The tyrphostin AG1478 inhibits proliferation and induces death of liver tumor cells through EGF receptor-dependent and independent mechanisms. Biochemical pharmacology. 2011;82(11):1583-92.
- Levy L, Renard CA, Wei Y, Buendia MA. Genetic alterations and oncogenic pathways in hepatocellular carcinoma. Annals of the New York Academy of Sciences. 2002;963:21-36.
- Peng H, Begg GE, Schultz DC, Friedman JR, Jensen DE, Speicher DW, et al. Reconstitution of the KRAB-KAP-1 repressor complex: a model system for defining the molecular anatomy of RING-B box-coiled-coil domain-mediated protein-protein interactions. Journal of molecular biology. 2000;295(5):1139-62.
- Peng H, Feldman I, Rauscher FJ, 3rd. Hetero-oligomerization among the TIF family of RBCC/TRIM domain-containing nuclear cofactors: a potential mechanism for regulating the switch between coactivation and corepression. Journal of molecular biology. 2002;320(3):629-44.
- Chambon M, Orsetti B, Berthe ML, Bascoul-Mollevi C, Rodriguez C, Duong V, et al. Prognostic significance of TRIM24/TIF-1alpha gene expression in breast cancer. The American journal of pathology. 2011;178(4):1461-9.
- Ding ZY, Jin GN, Wang W, Chen WX, Wu YH, Ai X, et al. Reduced expression of transcriptional intermediary factor 1 gamma promotes metastasis and indicates poor prognosis of hepatocellular carcinoma. Hepatology (Baltimore, Md). 2014;60(5):1620-36.
- Jain AK, Allton K, Iacovino M, Mahen E, Milczarek RJ, Zwaka TP, et al. p53 regulates cell cycle and microRNAs to promote differentiation of human embryonic stem cells. PLoS biology. 2012;10(2):e1001268.
- Yang CK, Yu TD, Han CY, Qin W, Liao XW, Yu L, et al. Genome-Wide Association Study of MKI67 Expression and its Clinical Implications in HBV-Related Hepatocellular Carcinoma in Southern China. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology. 2017;42(4):1342-57.
- Liu J, Rao J, Lou X, Zhai J, Ni Z, Wang X. Upregulated TRIM11 Exerts its Oncogenic Effects in Hepatocellular Carcinoma Through Inhibition of P53. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology. 2017;44(1):255-66.
- Wang X, Shi W, Shi H, Lu S, Wang K, Sun C, et al. TRIM11 overexpression promotes proliferation, migration and invasion of lung cancer cells. Journal of experimental & clinical cancer research : CR. 2016;35(1):100.
- Morris SM, Baek JY, Koszarek A, Kanngurn S, Knoblaugh SE, Grady WM. Transforming growth factor-beta signaling promotes hepatocarcinogenesis induced by p53 loss. Hepatology (Baltimore, Md). 2012;55(1):121-31.
- Liu M, Zhang X, Cai J, Li Y, Luo Q, Wu H, et al. Downregulation of TRIM58 expression is associated with a poor patient outcome and enhances colorectal cancer cell invasion. Oncology reports. 2018;40(3):1251-60.
- Zou Z, Anisowicz A, Hendrix MJ, Thor A, Neveu M, Sheng S, et al. Maspin, a serpin with tumor-suppressing activity in human mammary epithelial cells. Science (New York, NY). 1994;263(5146):526-9.
- Trepo E, Romeo S, Zucman-Rossi J, Nahon P. PNPLA3 gene in liver diseases. Journal of hepatology. 2016;65(2):399-412.
- Rotman Y, Koh C, Zmuda JM, Kleiner DE, Liang TJ. The association of genetic variability in patatin-like phospholipase domain-containing protein 3 (PNPLA3) with histological severity of nonalcoholic fatty liver disease. Hepatology (Baltimore, Md). 2010;52(3):894-903.
- Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nature reviews Gastroenterology & hepatology. 2012;9(10):599-608.
- Cani PD. Human gut microbiome: hopes, threats and promises. Gut. 2018;67(9):1716-25.
- Aagaard K, Petrosino J, Keitel W, Watson M, Katancik J, Garcia N, et al. The Human Microbiome Project strategy for comprehensive sampling of the human microbiome and why it matters. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2013;27(3):1012-22.
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59-65.
- Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504(7480):446-50.
- Falony G, Vieira-Silva S, Raes J. Microbiology Meets Big Data: The Case of Gut Microbiota-Derived Trimethylamine. Annual review of microbiology. 2015;69:305-21.
- Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS biology. 2007;5(7):e177.
- Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107(26):11971-5.
- Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Science translational medicine. 2014;6(237):237ra65.
- Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, et al. The NIH Human Microbiome Project. Genome research. 2009;19(12):2317-23.
- Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207-14.
- Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157(1):121-41.
- The Integrative Human Microbiome Project: dynamic analysis of microbiome-host omics profiles during periods of human health and disease. Cell Host Microbe. 2014;16(3):276-89.
- Lloyd-Price J, Mahurkar A, Rahnavard G, Crabtree J, Orvis J, Hall AB, et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature. 2017;550(7674):61-6.
- Delgado AP, Brandao P, Chapado MJ, Hamid S, Narayanan R. Open reading frames associated with cancer in the dark matter of the human genome. Cancer genomics & proteomics. 2014;11(4):201-13.
- Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452(7189):872-6.
- Knight R, Callewaert C, Marotz C, Hyde ER, Debelius JW, McDonald D, et al. The Microbiome and Human Biology. Annual Review of Genomics and Human Genetics. 2017;18(1):65-86.
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444(7122):1022-3.
- Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452(7186):423-8.
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027-31.
- Hernandez-Pacheco N, Farzan N, Francis B, Karimi L, Repnik K, Vijverberg SJ, et al. Genome-wide association study of inhaled corticosteroid response in admixed children with asthma. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology. 2019;49(6):789-98.
- Garcia-Sanchez A, Isidoro-Garcia M, Garcia-Solaesa V, Sanz C, Hernandez-Hernandez L, Padron-Morales J, et al. Genome-wide association studies (GWAS) and their importance in asthma. Allergologia et immunopathologia. 2015;43(6):601-8.
- Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448(7152):470-3.
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104(34):13780-5.
- Mattson DL, Liang M. Hypertension: From GWAS to functional genomics-based precision medicine. Nature reviews Nephrology. 2017;13(4):195-6.
- Saeed M. Locus and gene-based GWAS meta-analysis identifies new diabetic nephropathy genes. Immunogenetics. 2018;70(6):347-53.
- Hakonarson H, Grant SF. Genome-wide association studies (GWAS): impact on elucidating the aetiology of diabetes. Diabetes/metabolism research and reviews. 2011;27(7):685-96.
- Hartmann P, Seebauer CT, Schnabl B. Alcoholic liver disease: the gut microbiome and liver cross talk. Alcoholism, clinical and experimental research. 2015;39(5):763-75.
- Simon-Sanchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, Berg D, et al. Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nature genetics. 2009;41(12):1308-12.
- Gamazon ER, Innocenti F, Wei R, Wang L, Zhang M, Mirkov S, et al. A genome-wide integrative study of microRNAs in human liver. BMC genomics. 2013;14:395.
- Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, et al. Gut Microbiota Regulate Motor Deficits and Neuroinflammation in a Model of Parkinson's Disease. Cell. 2016;167(6):1469-80.e12.
- Knights D, Lassen KG, Xavier RJ. Advances in inflammatory bowel disease pathogenesis: linking host genetics and the microbiome. Gut. 2013;62(10):1505-10.
- Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nature reviews Immunology. 2009;9(5):313-23.
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122(1):107-18.
- Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56(7):1761-72.
- Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome biology. 2012;13(9):R79.
- Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. 2014;15(3):382-92.
- Inan MS, Rasoulpour RJ, Yin L, Hubbard AK, Rosenberg DW, Giardina C. The luminal short-chain fatty acid butyrate modulates NF-kappaB activity in a human colonic epithelial cell line. Gastroenterology. 2000;118(4):724-34.
- Kreznar JH, Keller MP, Traeger LL, Rabaglia ME, Schueler KL, Stapleton DS, et al. Host Genotype and Gut Microbiome Modulate Insulin Secretion and Diet-Induced Metabolic Phenotypes. Cell reports. 2017;18(7):1739-50.
- Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci U S A. 2010;107(44):18933-8.
- Carmody RN, Gerber GK, Luevano JM, Jr., Gatti DM, Somes L, Svenson KL, et al. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe. 2015;17(1):72-84.
- Hanssen MJ, Hoeks J, Brans B, van der Lans AA, Schaart G, van den Driessche JJ, et al. Short-term cold acclimation improves insulin sensitivity in patients with type 2 diabetes mellitus. Nat Med. 2015;21(8):863-5.
- Chevalier C, Stojanovic O, Colin DJ, Suarez-Zamorano N, Tarallo V, Veyrat-Durebex C, et al. Gut Microbiota Orchestrates Energy Homeostasis during Cold. Cell. 2015;163(6):1360-74.
- Worthmann A, John C, Rühlemann MC, Baguhl M, Heinsen F-A, Schaltenberg N, et al. Cold-induced conversion of cholesterol to bile acids in mice shapes the gut microbiome and promotes adaptive thermogenesis. Nature Medicine. 2017;23:839.
- Zietak M, Kovatcheva-Datchary P, Markiewicz LH, Stahlman M, Kozak LP, Backhed F. Altered Microbiota Contributes to Reduced Diet-Induced Obesity upon Cold Exposure. Cell Metab. 2016;23(6):1216-23.
- Fujisaka S, Ussar S, Clish C, Devkota S, Dreyfuss JM, Sakaguchi M, et al. Antibiotic effects on gut microbiota and metabolism are host dependent. J Clin Invest. 2016;126(12):4430-43.
- Cho I, Yamanishi S, Cox L, Methe BA, Zavadil J, Li K, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488(7413):621-6.
- Maida A, Hansotia T, Longuet C, Seino Y, Drucker DJ. Differential importance of glucose-dependent insulinotropic polypeptide vs glucagon-like peptide 1 receptor signaling for beta cell survival in mice. Gastroenterology. 2009;137(6):2146-57.
- Ikeda K, Inoue S. TRIM proteins as RING finger E3 ubiquitin ligases. Advances in experimental medicine and biology. 2012;770:27-37.
- Hatakeyama S. TRIM proteins and cancer. Nature reviews Cancer. 2011;11(11):792-804.
- Kimura T, Jain A, Choi SW, Mandell MA, Schroder K, Johansen T, et al. TRIM-mediated precision autophagy targets cytoplasmic regulators of innate immunity. The Journal of cell biology. 2015;210(6):973-89.
- Lascano J, Uchil PD, Mothes W, Luban J. TRIM5 Retroviral Restriction Activity Correlates with the Ability To Induce Innate Immune Signaling. Journal of virology. 2016;90(1):308-16.
- Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of 'single protein RING finger' E3 ubiquitin ligases. BioEssays : news and reviews in molecular, cellular and developmental biology. 2005;27(11):1147-57.
- Moosmann P, Georgiev O, Le Douarin B, Bourquin JP, Schaffner W. Transcriptional repression by RING finger protein TIF1 beta that interacts with the KRAB repressor domain of KOX1. Nucleic acids research. 1996;24(24):4859-67.
- Russell DW. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem. 2003;72:137-74.
- Worthmann A, John C, Ruhlemann MC, Baguhl M, Heinsen FA, Schaltenberg N, et al. Cold-induced conversion of cholesterol to bile acids in mice shapes the gut microbiome and promotes adaptive thermogenesis. Nat Med. 2017;23(7):839-49.
- Kuipers F, Bloks VW, Groen AK. Beyond intestinal soap--bile acids in metabolic control. Nature reviews Endocrinology. 2014;10(8):488-98.
- Kuipers F, Groen AK. An unexpected role for bile acid synthesis in adaptation to low temperature. Nat Med. 2017;23(7):800-2.
- Postic C, Girard J. Contribution of de novo fatty acid synthesis to hepatic steatosis and insulin resistance: lessons from genetically engineered mice. J Clin Invest. 2008;118(3):829-38.
- Donnelly KL, Smith CI, Schwarzenberg SJ, Jessurun J, Boldt MD, Parks EJ. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. J Clin Invest. 2005;115(5):1343-51.
- Broeders EP, Nascimento EB, Havekes B, Brans B, Roumans KH, Tailleux A, et al. The Bile Acid Chenodeoxycholic Acid Increases Human Brown Adipose Tissue Activity. Cell Metab. 2015;22(3):418-26.
- Watanabe M, Houten SM, Mataki C, Christoffolete MA, Kim BW, Sato H, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439(7075):484-9.
- Lien F, Berthier A, Bouchaert E, Gheeraert C, Alexandre J, Porez G, et al. Metformin interferes with bile acid homeostasis through AMPK-FXR crosstalk. J Clin Invest. 2014;124(3):1037-51.
- Backhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci U S A. 2007;104(3):979-84.
- Yang F, Huang X, Yi T, Yen Y, Moore DD, Huang W. Spontaneous development of liver tumors in the absence of the bile acid receptor farnesoid X receptor. Cancer Res. 2007;67(3):863-7.
- Kim I, Morimura K, Shah Y, Yang Q, Ward JM, Gonzalez FJ. Spontaneous hepatocarcinogenesis in farnesoid X receptor-null mice. Carcinogenesis. 2007;28(5):940-6.
- Sayin SI, Wahlstrom A, Felin J, Jantti S, Marschall HU, Bamberg K, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17(2):225-35.
- Claus SP, Tsang TM, Wang Y, Cloarec O, Skordi E, Martin FP, et al. Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Molecular systems biology. 2008;4:219.
- Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nature reviews Drug discovery. 2008;7(8):678-93.
- Carey EJ, Ali AH, Lindor KD. Primary biliary cirrhosis. Lancet (London, England). 2015;386(10003):1565-75.
- Mobraten K, Haugbro T, Karlstrom E, Kleiveland CR, Lea T. Activation of the bile acid receptor TGR5 enhances LPS-induced inflammatory responses in a human monocytic cell line. Journal of receptor and signal transduction research. 2015;35(5):402-9.
- Inagaki T, Moschetta A, Lee YK, Peng L, Zhao G, Downes M, et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci U S A. 2006;103(10):3920-5.
- Shea HC, Head DD, Setchell KD, Russell DW. Analysis of HSD3B7 knockout mice reveals that a 3alpha-hydroxyl stereochemistry is required for bile acid function. Proc Natl Acad Sci U S A. 2007;104(28):11526-33.
- Groothuis GM, Hardonk MJ, Keulemans KP, Nieuwenhuis P, Meijer DK. Autoradiographic and kinetic demonstration of acinar heterogeneity of taurocholate transport. The American journal of physiology. 1982;243(6):G455-62.
- Shaib YH, El-Serag HB, Davila JA, Morgan R, McGlynn KA. Risk factors of intrahepatic cholangiocarcinoma in the United States: a case-control study. Gastroenterology. 2005;128(3):620-6.
- Wang Y, Aoki H, Yang J, Peng K, Liu R, Li X, et al. The role of sphingosine 1-phosphate receptor 2 in bile-acid-induced cholangiocyte proliferation and cholestasis-induced liver injury in mice. Hepatology (Baltimore, Md). 2017;65(6):2005-18.
- Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, Luk A, et al. Identification of a nuclear receptor for bile acids. Science (New York, NY). 1999;284(5418):1362-5.
- Duane WC, Schteingart CD, Ton-Nu HT, Hofmann AF. Validation of [22,23-3H]cholic acid as a stable tracer through conversion to deoxycholic acid in human subjects. Journal of lipid research. 1996;37(2):431-6.
- Pullinger CR, Eng C, Salen G, Shefer S, Batta AK, Erickson SK, et al. Human cholesterol 7alpha-hydroxylase (CYP7A1) deficiency has a hypercholesterolemic phenotype. J Clin Invest. 2002;110(1):109-17.
- Sirvent A, Verhoeven AJ, Jansen H, Kosykh V, Darteil RJ, Hum DW, et al. Farnesoid X receptor represses hepatic lipase gene expression. Journal of lipid research. 2004;45(11):2110-5.
- Corpechot C, Carrat F, Bahr A, Chretien Y, Poupon RE, Poupon R. The effect of ursodeoxycholic acid therapy on the natural course of primary biliary cirrhosis. Gastroenterology. 2005;128(2):297-303.
- Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev. 2009;89(1):147-91.
- Keitel V, Reinehr R, Gatsios P, Rupprecht C, Gorg B, Selbach O, et al. The G-protein coupled bile salt receptor TGR5 is expressed in liver sinusoidal endothelial cells. Hepatology (Baltimore, Md). 2007;45(3):695-704.
- Wang YD, Chen WD, Wang M, Yu D, Forman BM, Huang W. Farnesoid X receptor antagonizes nuclear factor kappaB in hepatic inflammatory response. Hepatology (Baltimore, Md). 2008;48(5):1632-43.
- Chen WD, Wang YD, Zhang L, Shiah S, Wang M, Yang F, et al. Farnesoid X receptor alleviates age-related proliferation defects in regenerating mouse livers by activating forkhead box m1b transcription. Hepatology (Baltimore, Md). 2010;51(3):953-62.
- Meng Z, Liu N, Fu X, Wang X, Wang YD, Chen WD, et al. Insufficient bile acid signaling impairs liver repair in CYP27(-/-) mice. Journal of hepatology. 2011;55(4):885-95.
- Huang W, Ma K, Zhang J, Qatanani M, Cuvillier J, Liu J, et al. Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science (New York, NY). 2006;312(5771):233-6.
- Ridlon JM, Harris SC, Bhowmik S, Kang DJ, Hylemon PB. Consequences of bile salt biotransformations by intestinal bacteria. Gut microbes. 2016;7(1):22-39.
- Schramm C. Bile Acids, the Microbiome, Immunity, and Liver Tumors. The New England journal of medicine. 2018;379(9):888-90.
- Medzhitov R, Janeway C, Jr. Innate immunity. The New England journal of medicine. 2000;343(5):338-44.
- Dunkelberger JR, Song WC. Complement and its role in innate and adaptive immune responses. Cell research. 2010;20(1):34-50.
- Brubaker SW, Bonham KS, Zanoni I, Kagan JC. Innate immune pattern recognition: a cell biological perspective. Annual review of immunology. 2015;33:257-90.
- Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nature immunology. 2010;11(5):373-84.
- Kawai T, Akira S. TLR signaling. Seminars in immunology. 2007;19(1):24-32.
- O'Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7(5):353-64.
- Gordon S. Pattern recognition receptors: doubling up for the innate immune response. Cell. 2002;111(7):927-30.
- Takeuchi O, Akira S. Genetic approaches to the study of Toll-like receptor function. Microbes and infection. 2002;4(9):887-95.
- Wiest R, Albillos A, Trauner M, Bajaj JS, Jalan R. Targeting the gut-liver axis in liver disease. Journal of hepatology. 2017;67(5):1084-103.
- Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science (New York, NY). 1998;282(5396):2085-8.
- Shalapour S, Karin M. Immunity, inflammation, and cancer: an eternal fight between good and evil. J Clin Invest. 2015;125(9):3347-55.
- Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol. 2009;9(10):692-703.
- Boutens L, Stienstra R. Adipose tissue macrophages: going off track during obesity. Diabetologia. 2016;59(5):879-94.
- Quigley EM. Gut bacteria in health and disease. Gastroenterol Hepatol (N Y). 2013;9(9):560-9.
- Kawai T, Akira S. The roles of TLRs, RLRs and NLRs in pathogen recognition. International immunology. 2009;21(4):317-37.
- Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449(7164):819-26.
- Macpherson AJ, Slack E. The functional interactions of commensal bacteria with intestinal secretory IgA. Current opinion in gastroenterology. 2007;23(6):673-8.
- Steinman RM. Decisions about dendritic cells: past, present, and future. Annual review of immunology. 2012;30:1-22.
- Singla B, Ghoshal P, Lin H, Wei Q, Dong Z, Csanyi G. PKCdelta-Mediated Nox2 Activation Promotes Fluid-Phase Pinocytosis of Antigens by Immature Dendritic Cells. Frontiers in immunology. 2018;9:537.
- Sarkar K, Kruhlak MJ, Erlandsen SL, Shaw S. Selective inhibition by rottlerin of macropinocytosis in monocyte-derived dendritic cells. Immunology. 2005;116(4):513-24.
- Weiner LM, Surana R, Wang S. Monoclonal antibodies: versatile platforms for cancer immunotherapy. Nat Rev Immunol. 2010;10(5):317-27.
- Ouyang FZ, Wu RQ, Wei Y, Liu RX, Yang D, Xiao X, et al. Dendritic cell-elicited B-cell activation fosters immune privilege via IL-10 signals in hepatocellular carcinoma. Nature communications. 2016;7:13453.
- Merger M, Viney JL, Borojevic R, Steele-Norwood D, Zhou P, Clark DA, et al. Defining the roles of perforin, Fas/FasL, and tumour necrosis factor alpha in T cell induced mucosal damage in the mouse intestine. Gut. 2002;51(2):155-63.
- Zhou M, Wen Z, Cheng F, Ma J, Li W, Ren H, et al. Tumor-released autophagosomes induce IL-10-producing B cells with suppressive activity on T lymphocytes via TLR2-MyD88-NF-kappaB signal pathway. Oncoimmunology. 2016;5(7):e1180485.
- Gao R, Ma J, Wen Z, Yang P, Zhao J, Xue M, et al. Tumor cell-released autophagosomes (TRAP) enhance apoptosis and immunosuppressive functions of neutrophils. Oncoimmunology. 2018;7(6):e1438108.
- Geering B, Simon HU. Peculiarities of cell death mechanisms in neutrophils. Cell death and differentiation. 2011;18(9):1457-69.
- Enders A, Bouillet P, Puthalakath H, Xu Y, Tarlinton DM, Strasser A. Loss of the pro-apoptotic BH3-only Bcl-2 family member Bim inhibits BCR stimulation-induced apoptosis and deletion of autoreactive B cells. The Journal of experimental medicine. 2003;198(7):1119-26.
- Shalapour S, Lin XJ, Bastian IN, Brain J, Burt AD, Aksenov AA, et al. Inflammation-induced IgA+ cells dismantle anti-liver cancer immunity. Nature. 2017;551(7680):340-5.
- Mittal D, Gubin MM, Schreiber RD, Smyth MJ. New insights into cancer immunoediting and its three component phases--elimination, equilibrium and escape. Current opinion in immunology. 2014;27:16-25.
- Mittal D, Young A, Stannard K, Yong M, Teng MW, Allard B, et al. Antimetastatic effects of blocking PD-1 and the adenosine A2A receptor. Cancer Res. 2014;74(14):3652-8.
- Marabelle A, Kohrt H, Sagiv-Barfi I, Ajami B, Axtell RC, Zhou G, et al. Depleting tumor-specific Tregs at a single site eradicates disseminated tumors. J Clin Invest. 2013;123(6):2447-63.
- Li Y, Hahn T, Garrison K, Cui ZH, Thorburn A, Thorburn J, et al. The vitamin E analogue alpha-TEA stimulates tumor autophagy and enhances antigen cross-presentation. Cancer Res. 2012;72(14):3535-45.
- Nair S, Boddupalli CS, Verma R, Liu J, Yang R, Pastores GM, et al. Type II NKT-TFH cells against Gaucher lipids regulate B-cell immunity and inflammation. Blood. 2015;125(8):1256-71.
- Bald T, Landsberg J, Lopez-Ramos D, Renn M, Glodde N, Jansen P, et al. Immune cell-poor melanomas benefit from PD-1 blockade after targeted type I IFN activation. Cancer discovery. 2014;4(6):674-87.
- Thomas DL, Seeff LB. Natural history of hepatitis C. Clinics in liver disease. 2005;9(3):383-98, vi.
- Hatakeyama S. TRIM Family Proteins: Roles in Autophagy, Immunity, and Carcinogenesis. Trends in biochemical sciences. 2017;42(4):297-311.
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nature reviews Molecular cell biology. 2009;10(7):458-67.
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147(4):728-41.
- Sooro MA, Zhang N, Zhang P. Targeting EGFR-mediated autophagy as a potential strategy for cancer therapy. International journal of cancer. 2018;143(9):2116-25.
- Liu Z, Xiao Y, Torresilla C, Rassart E, Barbeau B. Implication of Different HIV-1 Genes in the Modulation of Autophagy. Viruses. 2017;9(12).
- White E. Deconvoluting the context-dependent role for autophagy in cancer. Nature reviews Cancer. 2012;12(6):401-10.
- Weiner LM, Lotze MT. Tumor-cell death, autophagy, and immunity. The New England journal of medicine. 2012;366(12):1156-8.
- Sakai Y, Koller A, Rangell LK, Keller GA, Subramani S. Peroxisome degradation by microautophagy in Pichia pastoris: identification of specific steps and morphological intermediates. The Journal of cell biology. 1998;141(3):625-36.
- Wen ZF, Liu H, Gao R, Zhou M, Ma J, Zhang Y, et al. Tumor cell-released autophagosomes (TRAPs) promote immunosuppression through induction of M2-like macrophages with increased expression of PD-L1. Journal for immunotherapy of cancer. 2018;6(1):151.
- Yuan W, Tuttle DL, Shi YJ, Ralph GS, Dunn WA, Jr. Glucose-induced microautophagy in Pichia pastoris requires the alpha-subunit of phosphofructokinase. Journal of cell science. 1997;110 ( Pt 16):1935-45.
- Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, et al. The tripartite motif family identifies cell compartments. The EMBO journal. 2001;20(9):2140-51.
- Huang Y, Yu Y, Yang Y, Yang M, Zhou L, Huang X, et al. Fish TRIM8 exerts antiviral roles through regulation of the proinflammatory factors and interferon signaling. Fish & shellfish immunology. 2016;54:435-44.
- Huang Y, Yang M, Yu Y, Yang Y, Zhou L, Huang X, et al. Grouper TRIM13 exerts negative regulation of antiviral immune response against nodavirus. Fish & shellfish immunology. 2016;55:106-15.
- Shafritz DA, Dabeva MD. Liver stem cells and model systems for liver repopulation. Journal of hepatology. 2002;36(4):552-64.
- Sell S. Heterogeneity and plasticity of hepatocyte lineage cells. Hepatology (Baltimore, Md). 2001;33(3):738-50.
- Fausto N. Liver regeneration and repair: hepatocytes, progenitor cells, and stem cells. Hepatology (Baltimore, Md). 2004;39(6):1477-87.
- Knight B, Yeoh GC, Husk KL, Ly T, Abraham LJ, Yu C, et al. Impaired preneoplastic changes and liver tumor formation in tumor necrosis factor receptor type 1 knockout mice. The Journal of experimental medicine. 2000;192(12):1809-18.
- Fausto N, Campbell JS, Riehle KJ. Liver regeneration. Hepatology (Baltimore, Md). 2006;43(2 Suppl 1):S45-53.
- Petersen BE, Grossbard B, Hatch H, Pi L, Deng J, Scott EW. Mouse A6-positive hepatic oval cells also express several hematopoietic stem cell markers. Hepatology (Baltimore, Md). 2003;37(3):632-40.
- Thorgeirsson SS, Grisham JW. Hematopoietic cells as hepatocyte stem cells: a critical review of the evidence. Hepatology (Baltimore, Md). 2006;43(1):2-8.
- Tewari M, Dobrzanski P, Mohn KL, Cressman DE, Hsu JC, Bravo R, et al. Rapid induction in regenerating liver of RL/IF-1 (an I kappa B that inhibits NF-kappa B, RelB-p50, and c-Rel-p50) and PHF, a novel kappa B site-binding complex. Molecular and cellular biology. 1992;12(6):2898-908.
- Cressman DE, Diamond RH, Taub R. Rapid activation of the Stat3 transcription complex in liver regeneration. Hepatology (Baltimore, Md). 1995;21(5):1443-9.
- Diehl AM, Yang SQ. Regenerative changes in C/EBP alpha and C/EBP beta expression modulate binding to the C/EBP site in the c-fos promoter. Hepatology (Baltimore, Md). 1994;19(2):447-56.
- Fausto N. Liver regeneration. Journal of hepatology. 2000;32(1 Suppl):19-31.
- Van Kaer L. alpha-Galactosylceramide therapy for autoimmune diseases: prospects and obstacles. Nat Rev Immunol. 2005;5(1):31-42.
- Schuster IS, Coudert JD, Andoniou CE, Degli-Esposti MA. "Natural Regulators": NK Cells as Modulators of T Cell Immunity. Frontiers in immunology. 2016;7:235.
- Exley M, Garcia J, Wilson SB, Spada F, Gerdes D, Tahir SM, et al. CD1d structure and regulation on human thymocytes, peripheral blood T cells, B cells and monocytes. Immunology. 2000;100(1):37-47.
- Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nature immunology. 2008;9(5):503-10.
- Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nature immunology. 2008;9(5):495-502.
- Zhou J, Peng H, Li K, Qu K, Wang B, Wu Y, et al. Liver-Resident NK Cells Control Antiviral Activity of Hepatic T Cells via the PD-1-PD-L1 Axis. Immunity. 2019;50(2):403-17.e4.
- Schildberg FA, Klein SR, Freeman GJ, Sharpe AH. Coinhibitory Pathways in the B7-CD28 Ligand-Receptor Family. Immunity. 2016;44(5):955-72.
- Jenne CN, Kubes P. Immune surveillance by the liver. Nature immunology. 2013;14(10):996-1006.
- Schrumpf E, Tan C, Karlsen TH, Sponheim J, Bjorkstrom NK, Sundnes O, et al. The biliary epithelium presents antigens to and activates natural killer T cells. Hepatology (Baltimore, Md). 2015;62(4):1249-59.
- Farin HF, Karthaus WR, Kujala P, Rakhshandehroo M, Schwank G, Vries RG, et al. Paneth cell extrusion and release of antimicrobial products is directly controlled by immune cell-derived IFN-gamma. The Journal of experimental medicine. 2014;211(7):1393-405.
- Zhang L, Yu X, Zheng L, Zhang Y, Li Y, Fang Q, et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature. 2018;564(7735):268-72.
- Biton M, Haber AL, Rogel N, Burgin G, Beyaz S, Schnell A, et al. T Helper Cell Cytokines Modulate Intestinal Stem Cell Renewal and Differentiation. Cell. 2018;175(5):1307-20.e22.
- Jadhav U, Saxena M, O'Neill NK, Saadatpour A, Yuan GC, Herbert Z, et al. Dynamic Reorganization of Chromatin Accessibility Signatures during Dedifferentiation of Secretory Precursors into Lgr5+ Intestinal Stem Cells. Cell stem cell. 2017;21(1):65-77.e5.
- Yan KS, Gevaert O, Zheng GXY, Anchang B, Probert CS, Larkin KA, et al. Intestinal Enteroendocrine Lineage Cells Possess Homeostatic and Injury-Inducible Stem Cell Activity. Cell stem cell. 2017;21(1):78-90.e6.
- Joyce JA, Fearon DT. T cell exclusion, immune privilege, and the tumor microenvironment. Science (New York, NY). 2015;348(6230):74-80.
- Segal NH, Parsons DW, Peggs KS, Velculescu V, Kinzler KW, Vogelstein B, et al. Epitope landscape in breast and colorectal cancer. Cancer Res. 2008;68(3):889-92.
- Cao W, Kayama H, Chen ML, Delmas A, Sun A, Kim SY, et al. The Xenobiotic Transporter Mdr1 Enforces T Cell Homeostasis in the Presence of Intestinal Bile Acids. Immunity. 2017;47(6):1182-96.e10.
- Pavlidis P, Heneghan M, Hayee B. Cholestyramine treats primary sclerosing cholangitis-associated inflammatory bowel disease. Journal of Crohn's & colitis. 2015;9(2):210.
- Lindner S, Dahlke K, Sontheimer K, Hagn M, Kaltenmeier C, Barth TF, et al. Interleukin 21-induced granzyme B-expressing B cells infiltrate tumors and regulate T cells. Cancer Res. 2013;73(8):2468-79.
- Lampropoulou V, Calderon-Gomez E, Roch T, Neves P, Shen P, Stervbo U, et al. Suppressive functions of activated B cells in autoimmune diseases reveal the dual roles of Toll-like receptors in immunity. Immunological reviews. 2010;233(1):146-61.
- Shao Y, Lo CM, Ling CC, Liu XB, Ng KT, Chu AC, et al. Regulatory B cells accelerate hepatocellular carcinoma progression via CD40/CD154 signaling pathway. Cancer letters. 2014;355(2):264-72.
- Ren Z, Peng H, Fu YX. PD-1 Shapes B Cells as Evildoers in the Tumor Microenvironment. Cancer discovery. 2016;6(5):477-8.
- Xiao X, Lao XM, Chen MM, Liu RX, Wei Y, Ouyang FZ, et al. PD-1hi Identifies a Novel Regulatory B-cell Population in Human Hepatoma That Promotes Disease Progression. Cancer discovery. 2016;6(5):546-59.
- Peng D, Kryczek I, Nagarsheth N, Zhao L, Wei S, Wang W, et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature. 2015;527(7577):249-53.
- Liu RX, Wei Y, Zeng QH, Chan KW, Xiao X, Zhao XY, et al. Chemokine (C-X-C motif) receptor 3-positive B cells link interleukin-17 inflammation to protumorigenic macrophage polarization in human hepatocellular carcinoma. Hepatology (Baltimore, Md). 2015;62(6):1779-90.
- Faggioli F, Palagano E, Di Tommaso L, Donadon M, Marrella V, Recordati C, et al. B lymphocytes limit senescence-driven fibrosis resolution and favor hepatocarcinogenesis in mouse liver injury. Hepatology (Baltimore, Md). 2018;67(5):1970-85.
- Pitzalis C, Jones GW, Bombardieri M, Jones SA. Ectopic lymphoid-like structures in infection, cancer and autoimmunity. Nat Rev Immunol. 2014;14(7):447-62.
- Evans AT, Loeb KR, Shulman HM, Hassan S, Qiu WC, Hockenbery DM, et al. Fibrosing cholestatic hepatitis C after hematopoietic cell transplantation: report of 3 fatal cases. The American journal of surgical pathology. 2015;39(2):212-20.
- Masters JR. HeLa cells 50 years on: the good, the bad and the ugly. Nature reviews Cancer. 2002;2(4):315-9.
- Rahbari R, Sheahan T, Modes V, Collier P, Macfarlane C, Badge RM. A novel L1 retrotransposon marker for HeLa cell line identification. BioTechniques. 2009;46(4):277-84.
- Hirschfield GM, Liu X, Xu C, Lu Y, Xie G, Lu Y, et al. Primary biliary cirrhosis associated with HLA, IL12A, and IL12RB2 variants. The New England journal of medicine. 2009;360(24):2544-55.
- Webb GJ, Hirschfield GM. Using GWAS to identify genetic predisposition in hepatic autoimmunity. Journal of autoimmunity. 2016;66:25-39.
- Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science (New York, NY). 2004;305(5685):872-4.
Citation: Okoli IP (2019) Tripartite Motif Cofactors, a Novel Gene Target for Liver Cancer via Regulating the Immune Cells and Gut Microbiome: A Review. Biochem Anal Biochem 8:239. doi: 10.35248/2167-0889.19.8.239
Copyright: © 2019 Okoli IP. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.