PMC/PubMed Indexed Articles
Indexed In
- Academic Keys
- ResearchBible
- CiteFactor
- Access to Global Online Research in Agriculture (AGORA)
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Mini Review - (2021) Volume 10, Issue 2
The Development of in Vitro Tools for Understanding the Physiology of Human Peripheral Sensory Neurons
Yoshie Umehara1, Mitsutoshi Tominaga2,3, François Niyonsaba1,4 and Kenji Takamori2,3,5*2Department of Juntendo Itch Research Center, Institute for Environmental and Gender-Specific Medicine, Juntendo University Graduate School of Medicine, Chiba, Japan
3Department of Anti-Aging Skin Research Laboratory, Juntendo University Graduate School of Medicine, Chiba, Japan
4Department of International Liberal Arts, Juntendo University, Tokyo, Japan
5Department of Dermatology, Juntendo University Urayasu Hospital, Chiba, Japan
Received: 17-Mar-2021 Published: 07-Apr-2021, DOI: 10.35248/2329-6682.21.10.164
Abstract
Somatic sensations, such as itch, pain, temperature and touch, are mediated by peripheral sensory neurons, which have cell bodies in Dorsal Root Ganglia (DRG) or trigeminal ganglia. Understanding the fundamental mechanisms underlying the transmission of somatosensory information is important for the development of tissue regeneration, disease modeling and treatment strategies. Genetic engineering techniques have identified sensory stimuli-associated transmission molecules in animals, whereas ethical difficulties collecting peripheral neurons from human DRG have limited analysis in humans. Peripheral sensory neurons derived from human cell resources are required to investigate the biology and pathophysiology underlying somatic sensations in humans. This review describes recently developed methods and tools used in physiological studies of human sensory neurons, as well as the results of these investigations.
Keywords
Human sensory neuron; Iitch; Pain; Neural crest; Human pluripotent stem cell; Direct reprogramming
Introduction
Primary sensory neurons mediate somatosensory information, including itch, pain, and temperature, touch and body position, to the central nervous system. These neurons contain cell bodies in Dorsal Root Ganglia (DRG) or trigeminal ganglia, with these cell bodies differing in somal size, the expression of ion channels and receptors, innervation territories, and electrophysiological properties [1]. Understanding the fundamental mechanisms underlying the transmission of sensory stimuli is informative for the tissue regeneration, disease modeling and treatment strategies.
Signaling molecules and receptors involved in the transmission of sensory stimuli have been identified in genetically engineered rodents and other animals. These mechanisms, however, differ markedly in humans and rodents. Novel therapeutic agents found in rodents may not be effective in humans. Although detailed investigations of human DRG neurons are required, the collection and analysis of peripheral neurons from human DRGs are very difficult due to limitations in cell numbers and ethical considerations regarding invasive harvesting techniques. Therefore, human peripheral sensory neurons derived from human cell resources are used for functional analyses. In recent years, methods have been developed to induce human peripheral neurons from Human Pluripotent Stem Cells (hPSCs) or fibroblasts. These approaches are useful for modeling pathologic conditions in humans and for designing therapeutic strategies. In addition, these approaches enable analysis of interactions between human sensory neurons and non-neuronal cells, such as immune cells, in coculture systems. This review highlights recent knowledge regarding the studies on the induction of human peripheral sensory neurons from hPSCs and human fibroblasts, which is essential for understanding somatosensory physiology in humans.
Development of peripheral sensory neurons
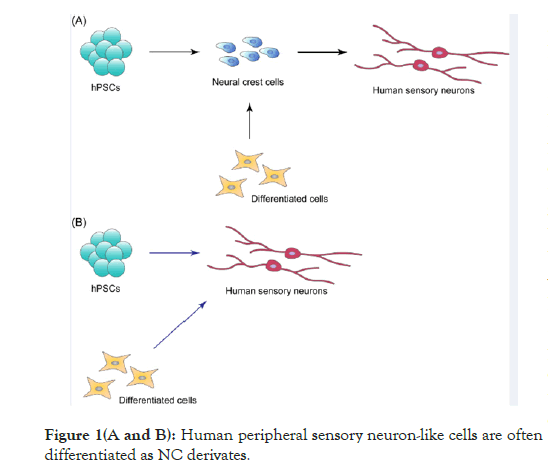
Peripheral sensory neurons are derived from a multipotent and migratory cell population, called neural crest (NC), which emerges at the interface between neural and non-neural ectoderm. NC differentiates into a wide variety of cell types, including neurons, glial cells, cranial bone, cartilage, smooth muscle and melanocytes, indicating that NC derivatives contribute to all tissues and organs at both the cephalic and trunk levels [2,3]. Deficiencies in NC development are therefore responsible for neurocristopathies, including Hirschsprung’s disease, DiGeorge syndrome (22q11.2 deletion syndrome), Waardenburg syndrome, CHARGE syndrome, Charcot-Marie-tooth disease, familial dysautonomia, congenital insensitivity to pain with anhidrosis, and pediatric cancers such as neuroblastoma [2,4]. Studies analyzing the pathology of neurocristopathies utilize human NC cells induced from hPSCs, including human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs) [5]. Human peripheral sensory neuron-like cells are often differentiated as NC derivates (Figure 1A). In addition, NC cells can be directly induced from human differentiated cells, including fibroblasts and blood cells. Human peripheral sensory neurons can also be generated by direct reprogramming of hPSCs and human differentiated cells (Figure 1B).
Induction of human peripheral sensory neurons through NC specification
No common method is presently available to induce NC from hPSCs. It is partially responsible for variations in NC markers, including transcription factors, receptors and carbohydrate chain [6]. Moreover, human NC has not been completely characterized. Most NC markers have been validated in animals, including mouse, chick, frog and lamprey, with few studies to date assessing the expression of some of these markers in human embryo [4,6,7]. To sort NC population, neurotrophin receptor p75 and carbohydrate chain HNK-1 are often used as surface antigen [6]. For example, p75+HNK-1+ NC cells could be efficiently (about 93%) generated from hESCs by blocking signaling of bone morphogenetic protein (BMP) and transforming growth factor (TGF)-β for 8 days, with the hESC-derived NC cells differentiating toward mesenchymal stem cells and neurons [8]. Moreover, blocking of BMP and TGF-β signaling in four hiPSC lines for 10 days generated 93% p75+HNK-1+ NC cells, resulting in the efficient differentiation of peripheral neurons from hiPSC-derived NC cells [9]. Inhibition of TGF-β and glycogen synthase kinase (GSK) 3β signaling for 7 days in three hESC lines and two hiPSC lines induced NC that expressed only p75, with induction efficiencies ranging from 70–80% [10]. These p75+ NC were found to differentiate toward multiple cell lineages, including mesenchymal stem cells and peripheral neurons [10]. The inhibition of BMP and GSK3β in three hESC and two hiPSC lines has also been reported to induce NC that expressed both p75 and HNK-1 with 70–90% induction efficiency, followed by the differentiation of peripheral neurons and mesenchymal stem cells from the NC cells [11]. SRY-box transcription factor (SOX) 10 has been detected in human, mouse and chick NC, which is used as a reporter gene for NC populations in the following studies. The inhibition of BMP, TGF-β and GSK3 in a SOX10::GFP reporter hESC line induced hESC-derived NC cells, with GFP expression peaking at 60% on day 11 [12]. Nociceptors (noxious stimulus–detecting neurons) and mature melanocytes were generated from the SOX10::GFP+ NC population [12,13]. More than 60% of SOX10::GFP+ NC-induced neurons expressed tyrosine receptor kinase (TRK) A, which is encoded by the neurotrophic tyrosine kinase receptor (NTRK)-1 gene. In contrast, NTRK2 and NTRK3 were not detected, suggesting that GFP-positive NC population had high potency to differentiate into TRKA+ nociceptors [13]. However, almost all SOX10::GFP-positive and SOX10::GFP-negative cell populations expressed p75, indicating that there are complicated distinct NC subtypes [13]. Indeed, almost all cells expressed p75 until day 6 of NC induction, HNK-1 expression increased through day 10 and SOX10 expression was maximal on day 8 in our study [9], suggesting that the times required for NC markers, such as p75, HNK-1 and SOX10, to reach maximal expression were different. These methods and NC induction efficiency in several previous studies could not be compared due to discordances in NC markers and assessment methods (Table 1). Comprehensive studies focusing on the diversity of NC subpopulations and differentiation potency would be necessary for establishment of a common method.
| Cell source | NC induction method | NC markers | Characterization of sensory neuron | Reference |
|---|---|---|---|---|
| hESC line (H1) | Blockade of BMP and TGF-β | p75 and HNK1 (Flow cytometry) TFAP2A (Immunostaining) TFAP2A , p75 , PAX3, SOX9, BRN3A (POU4F1) (qPCR) | None | 8 |
| iPSC lines (201B7, iPS-TIG120–3f7, iPS-TIG107–4f1, iPS-TIG114–4f) | Blockade of BMP and TGF-β | p75 and HNK1 (Flow cytometry) SOX10 (Immunostaining) TFAP2A , SOX10 , SOX9 , SNAI1 , SNAI2 (qPCR) | Calcium responsiveness to histamine, BAM8-22, IL-4, IL-31, capsaicin and allyl isothiocyanate Expression of BRN3A and peripherin (Immunostaining) | 9 |
| hESC lines (KhES1, KhES3, H9) hiPSC lines (201B7, 414C2) ; | Inhibition of TGF-β and GSK3β | p75 (Flow cytometry) p75, TFAP2A (Immunostaining) SOX10 , TWIST , TFAP2A (qPCR) | None | 10 |
| hESC lines (WA09, RUES1, RUES2) | Inhibition of TGF-β and GSK3β | p75 and HNK1 (Flow cytometry) p75, HNK1, TFAP2A (Immunostaining) p75, SOX9, SOX10 , BRN3A, TFAP2A (qPCR) | None | 11 |
| hiPSC lines (Fib2- iPS4, Fib2-iPS5) | ||||
| hESC lines (WA09, SHEF1) hiPSC lines (C14, C72) ; | Inhibition of BMP, TGF-β and GSK3β | SOX10 (SOX10::GFP reporter) p75, HNK1 (Immunostaining) | Calcium responsiveness to capsaicin and α,β me-ATP | 12,13 |
| TRKA+ nociceptor (more than 60% of all cells measured by FACS) | ||||
| Expression of BRN3A, ISL1, Substance P, CGRP (Immunostaining) |
Note: qPCR, quantitative real-time PCR
Table 1: Studies for inducing human peripheral sensory neurons through NC specification.
Direct reprogramming of human peripheral sensory neurons
Few studies have been reported that several transcription factors generate functional peripheral sensory neurons from human fibroblasts (Table 2). Five transcription factors were found to convert mouse and human fibroblasts into nociceptors, with reprogramming efficiency being lower for human than for mouse nociceptors [14]. Human sensory neurons can be reprogrammed from fibroblasts by transient expression of just two transcription factors [15]. Although direct reprogramming of peripheral sensory neurons is relatively inefficient yet, these methods enable investigations of the pharmacology of human peripheral neurons.
| Cell source | Sensory neuron induction method | Characterization of sensory neuron | Reference |
|---|---|---|---|
| Human fibroblasts from familial dysautonomia patients and healthy controls | Direct reprograming by five transcription factors (Ascl1 , Myt1l , Ngn1 , Isl2 , Klf7 ) | Expression of peripherin (Immunostaining) | 14 |
| Human embryonic fibroblasts from iPSC and adult human dermal fibroblasts | Direct reprograming by coexpression of BRN3A with either Ngn1 or Ngn2 | Calcium responsiveness to capsaicin, menthol, mustard oil, histamine, chloroquine, BAM8-22 and SLIGRLExpression of TRKA, TRKB, TRKC, ISL1, peripherin, P75 (Immunostaining) | 15 |
Table 2: Direct reprogramming methods to human peripheral sensory neurons.
Functional analyses of human peripheral sensory neurons
Various molecules that function in peripheral sensations have been identified; these include intracellular signaling molecules, ion channels, G Protein-Coupled Receptors (GPCRs), tyrosine kinase receptors and transducer molecules. Transient Receptor Potential (TRP) ion channels, especially transient receptor potential vanilloid 1 (TRPV1) and transient receptor potential ankyrin 1 (TRPA1), are involved in the generation in mammals of peripheral sensations such as warmth, pain and itch [16]. TRP channels are activated by various stimuli, such as extracellular pH, heat, cold, adenosine triphosphate (ATP), capsaicin, allyl isothiocyanate and GPCR ligands and associated with the pathogenesis of disorders in peripheral sensation [17]. Functional analyses require induced human peripheral neurons to respond to biological, biochemical and physiological stimuli. The rapid progress of these investigations over the last decade has enhanced understanding of the biology and pathology associated with human sensory neurons.
SOX10::GFP+ NC-induced neurons have shown calcium responses associated with noxious stimuli, such as capsaicin and α,β-methylene-adenosine triphosphate (α,β me-ATP), which are agonists of TRPV1 and P2X7 receptor respectively [13]. Some itch-related stimuli, including histamine, BAM8-22, interleukin (IL)-4, IL-31, capsaicin and allyl isothiocyanate, were found to induce calcium responses in human peripheral neurons derived from p75+HNK-1+ NC cells [9]. In addition, a distinct inhibitor used for the induction of hiPSC-derived NC cells was found to affect the potency of peripheral neuronal differentiation [9]. Peripheral sensory neurons directly reprogrammed from human fibroblasts exhibited the functional and physiological properties of mature sensory neurons [15]. These neurons expressed TRKA, TRKB and TRKC in almost the same proportion (approximately 30%) and responded to various compounds, such as capsaicin, menthol, mustard oil, histamine, chloroquine, and the peptides BAM8-22 and SLIGRL-NH2.
Another study compared human peripheral neurons reprogrammed from fibroblasts of a subject with familial dysautonomia and a healthy control individual [14]. Compared with neurons derived from a healthy control subject, the patient-derived neurons showed reductions in the number of neuron-specific class III β-tubulin (Tuj1)-positive neurons, neurite outgrowth and the number of branches [14]. Peripheral neurons derived from subject-specific iPSCs could be used to model interindividual differences in pain sensation in a family containing subjects with inherited erythromelalgia, a well characterized human genetic model of chronic pain [18]. This study found that sensory neuron excitability and pain sensation different among individuals and that these differences were associated with gene variants [18].
Prospects in basic research and beyond
The induction of human peripheral neurons is a recently available technology that can be used to investigate peripheral sensation; on the one hand, the specification of induced peripheral neurons was not sufficient to clarify diverse sensations. Furthermore, the assortment and functional analyses of induced NC still remain the issues. Although global genetic profiles in induced cell populations have been analyzed frequently, these cell populations have not been characterized in detail. Quantitative polymerase chain reaction analysis of single cells at five time points during hESC differentiation into sensory neurons provided precise information on cellular heterogeneity within a culture [19]. Because many methods are used to induce NC, investigation of NC subtypes differing by expression different markers or differentiation potential by using genetic techniques may have benefit in the study of neurocristopathies. Similar to NC induction, gene reporter systems will be needed to determine optimal conditions for peripheral neuronal differentiation or the induction of specific populations such as nociceptors. Investigating specifications in human NC and peripheral neuron subpopulations is also important in studying the developmental biology in humans.
Future investigations of human peripheral sensory neurons may reveal the molecular biology of human sensations. These approaches are useful in modeling human pathologies and in the development of new treatments for abnormal sensations, including itch and pain.
Conclusion
In conclusion, novel approaches generating human peripheral sensory neurons have enable functional analyses, overcoming the ethical difficulties involved in collecting peripheral neurons from human DRGs, thus limiting the analysis of somatosensory biology in humans. These expanded findings may lead to advances in the study of somatosensory physiology and may be helpful for disease modeling, drug screening and tissue regeneration.
Acknowledgments
This work was partly supported by JSPS KAKENHI Grant number 26860386, a grant of Strategic Research Foundation Grant-aided Project for Private Universities from MEXT (Grant number S1311011), a grant of Strategic Research Foundation Grant-aided Project for Private Universities from Ministry of Education, Culture, Sport, Science, and Technology, Japan (MEXT), 2014-2018 (S1411007) and the Atopy (Allergy) Research Center, Juntendo University, Tokyo, Japan.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- Ikoma A, Steinhoff M, Ständer S. yosipovitch G, Schmelz M. Neurobiology of pruritus. Nat Rev Neurosci. 2006;7:535-547.
- Lee G, Chambers SM, Tomishima MJ, Studer L. Derivation of neural crest cells from human pluripotent stem cells. Nat Protoc. 2010;5(4):688.
- Dupin E, Calloni GW, Coelho-Aguiar JM, Le Douarin NM. The issue of the multipotency of the neural crest cells. Dev Biol. 2018;444 Suppl 1:S47-S59.
- Etchevers HC, Dupin E, Le Douarin NM. The diverse neural crest: From embryology to human pathology. Development. 2019;146(5).
- Srinivasan A, Toh YC. Human pluripotent stem cell-derived neural crest cells for tissue regeneration and disease modeling. Front Mol Neurosci. 2019;12:39.
- Milet C, Monsoro-Burq AH. Embryonic stem cell strategies to explore neural crest development in human embryos. Dev Biol. 2012;366(1):96-99.
- Betters E, Liu Y, Kjaeldgaard A, Sundström E, García-Castro MI. Analysis of early human neural crest development. Dev Biol. 2010;344(2):578-592.
- Zhu Q, Li M, Yan C, Lu Q, Wei S, Gao R, et al. Directed differentiation of human embryonic stem cells to neural crest stem cells, functional peripheral neurons, and corneal Keratocytes. Biotechnol J. 2017;12(12).
- Umehara Y, Toyama S, Tominaga M, Matsuda H, Takahashi N, Kamata Y, et al. Robust induction of neural crest cells to derive peripheral sensory neurons from human induced pluripotent stem cells. Sci Rep. 2020;10(1):4360.
- Fukuta M, Nakai Y, Kirino K, Nakagawa M, Sekiguchi K, Nagata S, et al. Derivation of mesenchymal stromal cells from pluripotent stem cells through a neural crest lineage using small molecule compounds with defined media. PLoS One. 2014;9(12):e112291.
- Menendez L, Yatskievych TA, Antin PB, Dalton S. Wnt signaling and a Smad pathway blockade direct the differentiation of human pluripotent stem cells to multipotent neural crest cells. Proc Natl Acad Sci U S A. 2011;108(48):19240-19245.
- Mica Y, Lee G, Chambers SM, Tomishima MJ, Studer L. Modeling neural crest induction, melanocyte specification, and disease-related pigmentation defects in hESCs and patient-specific iPSCs. Cell Rep. 2013;3(4):1140-1152.
- Chambers SM, Qi Y, Mica Y, Lee G, Zhang XJ, Niu L, et al. Combined small-molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nat Biotechnol. 2012;30(7):715-720.
- Wainger BJ, Buttermore ED, Oliveira JT, Mellin C, Lee S, Saber WA, et al. Modeling pain in vitro using nociceptor neurons reprogrammed from fibroblasts. Nat Neurosci. 2015;18(1):17-24.
- Blanchard JW, Eade KT, Szucs A, Sardo VL, Tsunemoto RK, Williams D, et al. Selective conversion of fibroblasts into peripheral sensory neurons. Nat Neurosci. 2015;18(1):25-35.
- Sousaca-Valente J, Andreou AP, Urban L, Nagy I. Transient receptor potential ion channels in primary sensory neurons as targets for novel analgesics. Br J Pharmacol. 2014;171(10):2508-2527.
- Kittaka H, Tominaga M. The molecular and cellular mechanisms of itch and the involvement of TRP channels in the peripheral sensory nervous system and skin. Allergol Int. 2017;66(1):22-30.
- Mis MA, Yang Y, Tanaka BS, Gomis-Perez C, Liu S, Dib-Hajj F, et al. Resilience to pain: a peripheral component identified using induced pluripotent stem cells and dynamic clamp. J Neurosci. 2019;39(3):382-392.
- Young GT, Gutteridge A, Fox HD, Wilbrey AL, Cao L, Cho LT, et al. Characterizing human stem cell–derived sensory neurons at the single-cell level reveals their ion channel expression and utility in pain research. Mol Ther. 2014;22(8):1530-1543.
Citation: Umehara Y, Tominaga M, Niyonsaba F, Takamori K (2021) The Development of In Vitro Tools for Understanding the Physiology of Human Peripheral Sensory Neurons. Gene Technol. 10:164.
Copyright: © 2021 Umehara Y, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.