PMC/PubMed Indexed Articles
Indexed In
- Open J Gate
- Academic Keys
- JournalTOCs
- ResearchBible
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research - (2019) Volume 7, Issue 3
POOL-SEQ Study of Bulgarian Centenarians Highlights the Relevance for Human Longevity of Gene Expression Pathways
Dimitar Serbezov1, Lubomir Balabanski1, Sena Karachanak-Yankova1, Radoslava Vazharova1,2, Desislava Nesheva1, Zora Hammoudeh1, Rada Staneva1, Marta Mihaylova1, Vera Damyanova1, Olga Antonova1, Dragomira Nikolova1, Savina Hadjidekova1 and Draga Toncheva1*2Department of Biology, Medicine Genetics and Microbiology, Sofia University, Sofia, Bulgaria
Received: 02-Aug-2019 Published: 23-Aug-2019
Abstract
In human longevity studies a large number of genetic variants with small effects have been identified, but these are not easily replicable in different populations. We have performed whole-exome sequencing of two DNA pools from: 32 Bulgarian centenarians and 61 young healthy controls, respectively. A total of 59935 filtered variants were discovered, 216 of which were included in Longevity Map database which lists 2843 longevity associated variants. Using Fisher’s exact test, 22 of these variants showed significantly higher allele frequency in the centenarian compared to the control pool and are thus positively associated with longevity. Other 24 variants had significantly higher frequency in the controls and could be considered as negatively associated with longevity. The risk C allele in rs429358 of the APOE gene was only detected in the control pool and with lower frequency compared to other populations. REACTOME analyses showed that over-represented pathways with positive longevity variants belong to expression/transcription network with leading role of TP53, interplaying with other genes (ATR, FANCD2, BAX, BRIP1), whereas those with negative longevity variants belong to the signal transduction network. Our results confirm the importance of studying centenarians in different populations to discover those combinations of variants that associate with longer health span.
Keywords
Centenarian; Exome; Molecular pathways
Introduction
Longevity is an exceedingly complex phenotype, a quantitative trait that depends on the cumulativethe cumulative actions of many genes and the environment. The genetic component that determines longevity has been estimated to be around 30% [1], and this genetic contribution increases with age. Centenarians and super-centenarians, the extreme phenotypes of human lifespan, are thus unique cohorts to study the genetic basis of longevity and the factors determining the risk of various agerelated disorders.
The genomics of multifactorial diseases or conditions require sequencing a large number of genomes at high coverage; this is mandatory both in order to reach sufficient power for case-control analysis and to compare the patterns of genetic variations across populations. Yet, few studies employing whole-exome or whole-genome sequencing in centenarian populations to identify risk or beneficial variants relevant to extreme longevity have been performed to date.
Genetic factors can contribute to longevity in at least two important ways: an individual may inherit certain genetic variants that confer disease resistance thereby prolonging lifespan, or may inherit genetic variants that predispose him or her to disease thus decreasing longevity. Human longevity studies have identified a large number of genetic variants; these however provide limited biological insights as they are of small effect and are not being easily replicated in different populations. One of the very few variants that has been associated with longevity across a number of studies is rs429358 in the APOE gene with lower frequency of C allele in centenarians [2]. Clarifying the role of such variants is further complicated by the evidence that they operate in intricate network of interactions through biological pathways to influence the final phenotype. New insights may thus come from the combined analysis of different genes or SNPs, especially when grouped by metabolic pathways or gene region. Pathway-based analyses thus allow the interpretation of variants with respect to the biological processes in which the affected genes and proteins are involved. As such, pathway analyses are an important follow-up for genomewide association studies (GWAS) to provide mechanistic insight about the underlying biology of longevity. By identifying well-annotated pathways that map to the lists of significant genes identified in a GWAS, biochemical hypotheses can be enumerated and tested. The effect on longevity of SNPs belonging to certain candidate pathways, such as the insulin/insulin like growth factor signaling (IIS) and DNA repair, have repeatedly been demonstrated [3,4]. Sequencing DNA pools (Pool-seq) is a cost-effective method widely used in genomic/exomic sequencing. Pool-seq methods have been repeatedly shown to provide reliable estimates of allele frequencies [5-7], making them particularly suitable for unravelling the genetic basis of complex traits such as longevity. In this study, we first obtain data on SNP and indel variants from whole-exome sequencing two DNA pools, one compiled with Bulgarian centenarians and the other with co-ethnic healthy young controls. Of all annotated variants, we select variants designated as being associated with longevity (LAVs) in the publicly accessible database Longevity Map [8]. The functional significance of the set of genes with variants that show significant allele frequency difference between the two pools was subsequently analyzed using REACTOME pathway platform.
Materials and Methods
Ethics statement, centenarian and control subjects recruitment
The project was approved by the Ethics committee of the Medical University of Sofia and was found to be in accordance with the requirements for conducting research with the participation of human subjects, as well as with the national and international legislation. Each participant in the study was informed with the aims of the project. Written informed consent was obtained from participants before collecting samples for DNA extraction. Centenarian health status and medical history for major age-related diseases were based on interviews conducted with the subjects. Tissue samples (saliva or blood sample) was collected from a group of Bulgarian centenarians [100 to 106 year old] and ethnically matched control group composed of young healthy individuals aged 25-30 years.
DNA isolation, WES sequencing and statistical analyses
DNA was extracted using QIAamp DNA Blood Mini Kit (Qiagen) and equimolar amounts of DNA were used to prepare two pools: one with 32 Bulgarian centenarians and one with 61 young individuals as controls. These were whole-exome sequenced using BGI v4 chemistry on a BGISEQ- 500 platform (by BGI Genomics) at a mean 250x coverage. Such high coverage is required for pool-seq sequencing to ensure that alleles with low frequency are also detected [5]. The obtained .VCF files were annotated using the web-based service wANNOVAR [9]. Following the ‘best practice’ recommendations for pool-seq data [7], we performed robust filtering on variant calling: genotype quality ≥ 99, mapping quality ≥ 60, number of reads per MAF>2, total depth of coverage above 30 and below 500.
The total number of variants annotated in both pools after applying these filters was 59935 (52870 SNPs and 7065 indels). The number of alleles reads for each variant in both pools was used to construct contingency tables and Fisher’s exact test was then deployed to evaluate the significance of the allele frequency differences. The allele frequency estimates obtained were compared with values taken from the publicly available resource for exome sequencing data, The Genome Aggregation Database (gnomAD) [10]. False Discovery Rate (FDR) adjustment of Benjamini and Hochberg [11] was used to reduce the number of false positives. All statistical analyses were performed using R scripts [12].
Longevity map
We used Longevity Map, a publicly accessible database of human genetic variants associated with longevity to compile a list of variants. This database lists a total of 2843 variants, 510 of which denoted to be significantly and 2333 denoted to be nonsignificantly associated with longevity. We then examined our pool-seq data for the presence and the frequency of these variants.
Pathway analysis
Pathway analysis methods use a variety of different strategies to aggregate or interpret individual marker or gene based phenotype association statistics to yield a single interpretable test statistic (or p-value) summarizing the strength of evidence of association between the pathway and the phenotype. Pathway analysis simplifies the interpretation by placing findings in context of prior knowledge, as well as the analysis (by reducing the multiple comparisons burden inherent to genomewide approaches). The functional significance of the assemblage of genes that have variants showing significant difference between the two pools was examined using the web-based platform REACTOME [13].
Results
Assessment of the accuracy of allele frequency estimation
In order to verify the adequacy of the population allele frequency estimation using poolseq data, we compared the estimated individual allele frequencies from the Bulgarian control group with allele frequencies from the gnomAD database. The obtained correlation coefficient (Pearson’s r) is 0.96, suggesting a high degree of accuracy of allele frequency estimation from pool-seq data.
Longevity associated variants (LAVs)
Of all 2843 variants designated to be associated with longevity in Longevity Map, 216 were discovered in both the centenarian and control pools. Forty six of these showed significant difference between the centenarian and control pools and they were in 43 genes.
Twenty two variants in 20 genes are positively associated with longevity (positive LAVs), i.e. their frequency is significantly higher in the centenarian pool (FDR adj. p-value<0.05) (Table 1).
| Chr (n) | Ref | Alt | Function | Gene | Exonic function | dbSNP | Frequency centenarians | Frequency controls | FDR P- value |
|---|---|---|---|---|---|---|---|---|---|
| Variants designated in LongevityMap as being significantly associated with longevity | |||||||||
| 17 | G | C | Exonic | TP53 | Nonsynonymous | rs1042522 | 0.844 | 0.724 | 0.001 |
| 7 | C | T | Exonic | EGFR | Synonymous | rs2072454 | 0.553 | 0.438 | 0.046 |
| 6 | A | G | ncRNA exonic | CMAHP | - | rs303006 | 0.854 | 0.705 | <0.0001 |
| Variants designated in LongevityMap as being non-significantly associated with longevity | |||||||||
| 2 | C | G | Exonic | CYP1B1 | Nonsynonymous | rs1056836 | 0.626 | 0.502 | 0.021 |
| 1 | T | C | Intronic | IL10 | - | rs1518111 | 0.741 | 0.565 | 0.005 |
| 11 | A | G | Exonic | GSTP1 | Nonsynonymous | rs1695 | 0.347 | 0.242 | 0.041 |
| 14 | G | A | Exonic | MLH3 | Nonsynonymous | rs175080 | 0.577 | 0.453 | 0.029 |
| 3 | C | T | Exonic | ATR | Synonymous | rs1802904 | 0.87 | 0.729 | <0.0001 |
| 19 | A | G | Intronic | BAX | - | rs1805419 | 0.839 | 0.663 | 0.001 |
| 11 | T | C | Exonic | ACTN3 | Unknown | rs1815739 | 0.766 | 0.599 | <0.0001 |
| 11 | T | C | Exonic | MCAM | Synonymous | rs2249466 | 0.802 | 0.689 | 0.013 |
| 3 | C | T | UTR3 | FANCD2 | - | rs3172417 | 0.39 | 0.283 | 0.013 |
| 5 | G | A | Intronic | RASA1 | - | rs3752862 | 0.588 | 0.387 | 0.048 |
| 8 | C | T | Intronic | RECQL4 | - | rs4251689 | 0.569 | 0.447 | 0.043 |
| 17 | C | G | Intronic | ACE | - | rs4295 | 0.783 | 0.557 | 0.013 |
| 17 | T | C | Intronic | ACE | - | rs4311 | 0.665 | 0.488 | 0.012 |
| 17 | A | G | Exonic | ACE | Synonymous | rs4331 | 0.529 | 0.351 | 0.009 |
| 17 | A | G | Exonic | BRIP1 | Nonsynonymous | rs4986764 | 0.585 | 0.472 | 0.018 |
| 11 | G | C | UTR3 | APOC3 | - | rs5128 | 0.925 | 0.847 | 0.011 |
| 17 | T | C | Exonic | SLC2A4 | Synonymous | rs5435 | 0.72 | 0.622 | 0.018 |
| 3 | G | A | Exonic | IMPG2 | Nonsynonymous | rs571391 | 0.633 | 0.48 | <0.0001 |
| 7 | C | T | Exonic | TAS2R16 | Nonsynonymous | rs860170 | 0.688 | 0.458 | <0.0001 |
| Chr: Chromosome Number; Ref: Referent Allele; Alt: Alternative Allele | |||||||||
Table 1: Longevity associated SNPs found in the pool-seq data from Bulgarian centenarians and a control group.
Three of these variants are designated as being significantly associated with longevity by Longevity Map, while the remaining 19 have been designated to be non-significantly associated.
Gene set analysis for molecular pathways based on genes with positive LAVs
Twelve out of 15 significant REACTOME pathways (FDR adj. p-value<0.05) with genes carrying positive LAVs are part of the gene expression/transcription network (GET) (Figure 1 and Table 2).
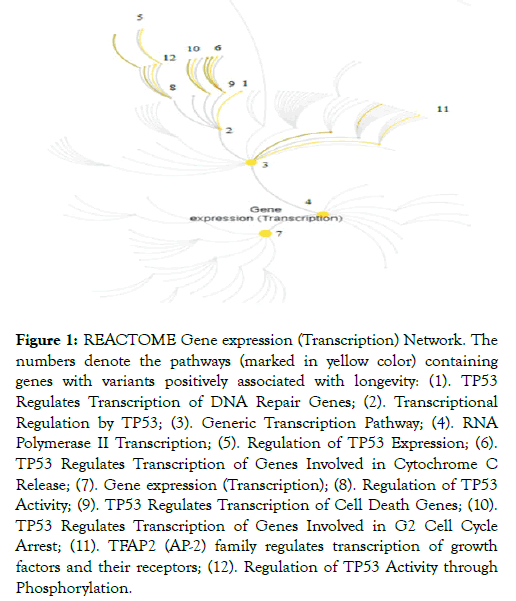
Figure 1: REACTOME Gene expression (Transcription) Network. The numbers denote the pathways (marked in yellow color) containing genes with variants positively associated with longevity: (1). TP53 Regulates Transcription of DNA Repair Genes; (2). Transcriptional Regulation by TP53; (3). Generic Transcription Pathway; (4). RNA Polymerase II Transcription; (5). Regulation of TP53 Expression; (6). TP53 Regulates Transcription of Genes Involved in Cytochrome C Release; (7). Gene expression (Transcription); (8). Regulation of TP53 Activity; (9). TP53 Regulates Transcription of Cell Death Genes; (10). TP53 Regulates Transcription of Genes Involved in G2 Cell Cycle Arrest; (11). TFAP2 (AP-2) family regulates transcription of growth factors and their receptors; (12). Regulation of TP53 Activity through Phosphorylation.
| Nr. | Pathway name | Entities found | Entities total | Entities ratio | Entities p-value | Entities FDR |
|---|---|---|---|---|---|---|
| Gene expression/Transcription | ||||||
| 1 | TP53 regulates transcription of DNA repair genes | 7 | 89 | 0.006 | 1.14E-09 | 2.68E-07 |
| Input: ATR, FANCD2, TP53 | ||||||
| 2 | Transcriptional regulation by TP53 | 11 | 486 | 0.034 | 5.16E-09 | 6.04E-07 |
| Input: ATR, BAX, BRIP1, FANCD2, TP53 | ||||||
| 3 | Generic transcription pathway | 13 | 1525 | 0.108 | 1.25E-05 | 9.75E-04 |
| Input: ATR, BAX, BRIP1, EGFR, FANCD2, TP53 | ||||||
| 4 | RNA polymerase II transcription | 13 | 1664 | 0.118 | 3.21E-05 | 1.85E-03 |
| Input: ATR, BAX, BRIP1, EGFR, FANCD2, TP53 | ||||||
| 5 | Regulation of TP53 expression | 2 | 4 | 0 | 4.00E-05 | 1.85E-03 |
| Input: TP53 | ||||||
| 6 | TP53 Regulates transcription of genes involved in Cytochrome C release | 3 | 33 | 0.002 | 6.00E-05 | 2.33E-03 |
| Input: BAX, TP53 | ||||||
| 7 | Gene expression (Transcription) | 13 | 1822 | 0.129 | 8.38E-05 | 2.77E-03 |
| Input: ATR, BAX, BRIP1, EGFR, FANCD2, TP53 | ||||||
| 8 | Regulation of TP53 activity | 4 | 178 | 0.013 | 6.79E-04 | 1.94E-02 |
| Input: ATR, BRIP1, TP53 | ||||||
| 9 | TP53 regulates transcription of cell death genes | 3 | 83 | 0.006 | 8.80E-04 | 1.94E-02 |
| Input: BAX, TP53 | ||||||
| 10 | TP53 Regulates Transcription of Genes involved in G2 cell cycle arrest | 2 | 21 | 0.001 | 1.06E-03 | 1.90E-02 |
| Input: BAX, TP53 | ||||||
| 11 | TFAP2 (AP-2) family regulates transcription of growth factors and their receptors | 2 | 21 | 0.001 | 1.06E-03 | 1.94E-02 |
| Input: EGFR | ||||||
| 12 | Regulation of TP53 activity through phosphorylation | 3 | 95 | 0.007 | 1.30E-03 | 1.94E-02 |
| Input: ATR | ||||||
| Cell cycle | ||||||
| 13 | G2/M DNA damage checkpoint | 3 | 81 | 0.006 | 8.21е-4 | 1.94е-2 |
| Input: ATR | ||||||
| Disease | ||||||
| 14 | Uptake and function of anthrax toxins | 2 | 22 | 0.002 | 1.16е-3 | 1.94е-2 |
| Input: ATR | ||||||
| Immune system | ||||||
| 15 | Interleukin-4 and Interleukin-13 signaling | 4 | 211 | 0.015 | 1.27е-3 | 1.94е-2 |
| Input: IL10, TP53 | ||||||
Table 2: Pathways, indicated by REACTOME to be significantly associated (FDR<1.94е-2) with the set of genes with LAVs.
Other significant pathways (FDR adj. p-value=1.94е-2) that REACTOME states to be constituted by genes from our positive LAVs gene set are: G2/M DNA damage checkpoint (part of the Cell cycle network); Interleukin-4 and Interleukin-13 signaling (part of the Disease network); uptake and function of anthrax toxins (part of the Immune system network).
Twenty four variants in 23 genes have significantly lower variant frequency in the centenarian pool and it can be speculated that these have disadvantageous effect on longevity (negative LAVs) (Table 3).
| Chr | Ref | Alt | Function | Gene | Exonic function | dbSNP | Frequency centenarians | Frequency controls | FDR p-value |
|---|---|---|---|---|---|---|---|---|---|
| Variants designated in LongevityMap as being significantly associated with longevity, with negative association in BG centenarians | |||||||||
| 14 | C | A | Intergenic | AKT1 | - | rs2498804 | 0.264 | 0.392 | 0.024 |
| 1 | A | G | Exonic | EXO1 | Nonsynonymous | rs735943 | 0.411 | 0.612 | 0.001 |
| Variants designated in LongevityMap as being non-significantly associated with longevity, with negative association in BG centenarians | |||||||||
| 16 | G | A | Intronic | PDPK1 | - | rs1005273 | 0.327 | 0.512 | 0.036 |
| 11 | A | G | Exonic | HRAS | Synonymous | rs12628 | 0.29 | 0.484 | <0.0001 |
| 15 | A | T | Exonic | PIF1 | Nonsynonymous | rs17802279 | 0.358 | 0.494 | 0.028 |
| 1 | T | G | Exonic | MTHFR | Nonsynonymous | rs1801131 | 0.28 | 0.396 | 0.017 |
| 1 | G | A | Exonic | MTHFR | Nonsynonymous | rs1801133 | 0.276 | 0.384 | 0.04 |
| 16 | A | G | Intronic | PRKCB | - | rs198145 | 0.209 | 0.373 | 0.003 |
| 7 | G | T | Exonic | TAS2R5 | Nonsynonymous | rs2227264 | 0.485 | 0.628 | 0.006 |
| 17 | G | A | Intronic | RPA1 | - | rs2270412 | 0.169 | 0.261 | 0.022 |
| 16 | C | G | Exonic | CLEC3A | Synonymous | rs2293776 | 0.213 | 0.334 | 0.006 |
| 19 | T | G | Exonic | ERCC2 | Synonymous | rs238406 | 0.419 | 0.559 | 0.023 |
| 2 | C | G | Intronic | MSH6 | - | rs3136367 | 0.619 | 0.773 | 0.001 |
| 19 | C | A | Exonic | CD3EAP | Nonsynonymous | rs3212986 | 0.183 | 0.438 | 0.001 |
| 16 | A | G | Exonic | IGFALS | Synonymous | rs3751893 | 0.688 | 0.812 | 0.004 |
| 15 | T | C | Intronic | BLM | - | rs3815003 | 0.201 | 0.287 | 0.041 |
| 3 | A | G | Intronic | ADCY5 | - | rs4482616 | 0.733 | 0.968 | <0.0001 |
| 7 | A | G | Exonic | IGFBP1 | Nonsynonymous | rs4619 | 0.151 | 0.347 | <0.0001 |
| 16 | T | G | ncRNA intronic | MT1JP | - | rs4784701 | 0.476 | 0.692 | <0.0001 |
| 1 | C | T | Exonic | NTRK1 | Synonymous | rs6337 | 0.676 | 0.785 | 0.02 |
| 2 | C | G | Intronic | XDH | - | rs761926 | 0.208 | 0.361 | <0.0001 |
| 7 | G | T | Intronic | NOS3 | - | rs7830 | 0.174 | 0.327 | 0.046 |
| 19 | G | C | Intronic | GPX4 | - | rs8178977 | 0.199 | 0.323 | <0.0001 |
| 2 | C | T | Exonic | FSHR | Nonsynonymous | rs6166 | 0.477 | 0.586 | 0.013 |
Table 3: Variants from LongevityMap with significantly lower frequency in BG centenarians.
Gene set analysis for molecular pathways based on genes with negative LAVs
Ten out of 20 significant REACTOME pathways (FDR adj. p-value<0.05) with genes carrying negative LAVs are members of the signal transduction network (Figure 2 and Table 4).
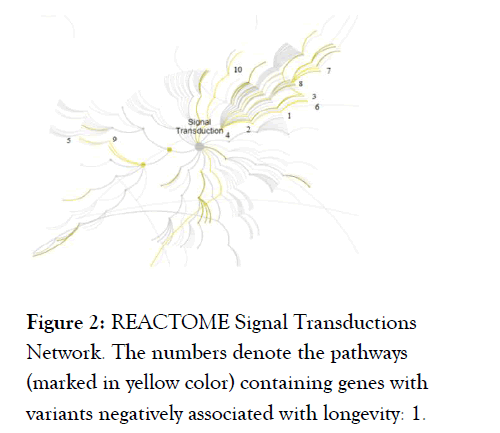
Figure 2. REACTOME Signal Transductions Network. The numbers denote the pathways (marked in yellow color) containing genes with variants negatively associated with longevity: 1.
| Nr. | Pathway name | Entities found | Entities total | Entities ratio | Entities p-value | Entities FDR |
|---|---|---|---|---|---|---|
| Signal transduction network | ||||||
| 1. | VEGFA-VEGFR2 Pathway Input: AKT1, HRAS, NOS3, PDPK1, PRKCB | 6 | 125 | 0.009 | 4.28е-7 | 1.41е-4 |
| 2. | Signaling by VEGF Input: AKT1, HRAS, NOS3, PDPK1, PRKCB | 6 | 134 | 0.009 | 6.4e-7 | 1.41e-4 |
| 3. | VEGFR2 mediated vascular permeability Input: AKT1, NOS3, PDPK1 | 4 | 44 | 0.003 | 3.55e-6 | 5.19e-4 |
| 4. | Signaling by receptor tyrosine kinases Input: AKT1, HRAS, NOS3, NTRK1, PDPK1, PRKCB | 8 | 521 | 0.037 | 2.05e-5 | 1.65e-3 |
| 5. | G beta:gamma signaling through PI3Kgamma Input: AKT1, PDPK1 | 3 | 29 | 0.002 | 4.48e-5 | 2.16e-3 |
| 6. | VEGFR2 mediated cell proliferation Input: HRAS, PDPK1, PRKCB | 3 | 30 | 0.002 | 5.00e-5 | 2.16e-3 |
| 7. | TRKA activation by NGF Input: NTRK1 | 2 | 5 | 0 | 6.54e-5 | 2.16e-3 |
| 8. | Signaling to ERKs Input: HRAS, NTRK1 | 3 | 39 | 0.003 | 1.07e-4 | 2.68e-3 |
| 9. | G-protein beta:gamma signaling Input: AKT1, PDPK1 | 3 | 39 | 0.003 | 1.07e-4 | 2.68e-3 |
| 10. | Non-genomic estrogen signaling Input: AKT1, HRAS, NOS3, PDPK1 | 4 | 109 | 0.008 | 1.20e-4 | 2.86e-3 |
| DNA repair | ||||||
| 11. | Mismatch repair (MMR) directed by MSH2:MSH6 (MutSalpha) Input: EXO, MSH6, RPA1 | 3 | 22 | 0.002 | 2,00e-05 | 1.65e-3 |
| 12. | Mismatch Repair Input: EXO, MSH6, RPA1 | 3 | 23 | 0.002 | 2.26e-5 | 1.65e-3 |
| 13. | HDR through single strand annealing (SSA) Input: BLM, EXO1, RPA1 | 3 | 39 | 0.003 | 1.07e-4 | 2.68e-3 |
| 14. | Presynaptic phase of homologous DNA pairing and strand exchange Input: BLM, EXO1, RPA1 | 3 | 41 | 0.003 | 1.24e-4 | 2.86e-3 |
| 15. | Homologous DNA pairing and strand exchange Input: BLM, EXO1, RPA1 | 3 | 44 | 0.003 | 1.53e-4 | 3.36e-3 |
| Immune system | ||||||
| 16. | CD28 dependent PI3K/Akt signaling Input: AKT1, PDPK1 | 3 | 26 | 0.002 | 3.24e-5 | 2.01e-3 |
| 17. | CD28 co-stimulation Input: AKT1, PDPK1 | 3 | 39 | 0.003 | 1.07e-4 | 2.68e-3 |
| Gene expression/Transcription | ||||||
| 18. | Regulation of TP53 activity Input: AKT1, BLM, EXO1, PDPK1, RPA1 | 5 | 178 | 0.013 | 5.56E-5 | 2.16e-3 |
| 19. | RNA polymerase I transcription termination Input: CD3EAP, ERCC2, RPA1 | 3 | 33 | 0.002 | 6.56e-5 | 2.16e-3 |
| Disease | ||||||
| 20. | Constitutive signaling by AKT1 E17K in cancer Input: AKT1, PDPK1 | 3 | 32 | 0.002 | 6.00e-5 | 2.16e-3 |
Table 4: Pathways, indicated by REACTOME to be significantly associated (FDR adj. p-value<1.94е-2) with the set of genes with variants negatively associated with longevity.
VEGFA-VEGFR2 Pathway; 2. Signaling by VEGF; 3. VEGFR2 mediated vascular permeability; 4. Signaling by Receptor Tyrosine Kinases; 5. G beta:gamma signaling through PI3Kgamma; 6.
VEGFR2 mediated cell proliferation; 7. TRKA activation by NGF; 8. Signaling to ERKs; 9. G-protein beta:gamma signaling; 10. Non-genomic estrogen signaling.
Other significant pathways that REACTOME indicates to be constituted by genes from this set are CD28 dependent PI3K/Akt signaling and CD28 costimulation (both part of the Immune system network); Regulation of TP53 activity and RNA polymerase I transcription termination (both part of the Gene expression/Transcription); Constitutive signaling by AKT1 E17K in cancer (Disease network).
The remaining 170 variants failed to replicate as a longevity-associated variants in Bulgarian centenarians as there is no significant differences in allele frequencies between the centenarian and control pools.
Discussion
Whole-exome sequencing on DNA pools from Bulgarian centenarians and a control group found 216 out of 2843 longevity associated variants (LAVs) listed in the Longevity Map database. Only 22 of these showed significantly higher allele frequency in the centenarian pool compared to the control pool and can thus be considered positively associated with longevity in Bulgarian population.
Longevity Map database designates only 3 of these 22 variants (in genes TP53, EGFR, CMAHP) to have significant association with longevity; while the remaining 19 variants, although determined to be non-significant LAVs, are significantly associated with longevity in Bulgarian population (see Table 1). The positive LAVs in our study are thus less than 1% of all variants listed in the Longevity Map database. A possible explanation for this low percentage is that longevity is a complex phenotype, being determined by interaction between genetic and environmental factors. LAVs should be studied in different populations as they must deal with different conditions and specific LAV combinations can determine longevity in different ethnic or geographical groups.
Pathway analysis of gene set with positive LAVs in Bulgarian population
Twelve out of 15 REACTOME overrepresented pathways in the set of genes with positive LAVs constitute the gene expression/transcription (GET) network (Figure 1). Part of the co-expressed genes, members of the same pathway, are controlled either by the same transcriptional factors or are functionally related, or are members of the same protein complex. A transcriptional regulatory network plays a major role in response to environmental influences and genetic disturbances and it consists of many components [14]. Although there has been much progress in establishing interactions between the molecular pathways in GET network, their role in centenarians remains unclear.
REACTOME pathways are hierarchically arranged, the top level pathway being “Gene expression/transcription” (Figure 1, pathway 7) in which participate the enzymes RNA polymerase I, II, III and mitochondrial RNA polymerase. It is followed by the pathways “RNA Polymerase II transcription” (Figure 1, pathway 4) and “Generic transcription pathway” in which cell- or tissuespecific regulation of differential gene transcription is mediated (Figure 1, pathway 3). These are fundamental pathways for gene expression and starting point for the other pathways in the network. The remaining significant pathways involve overlapping set of positive LAV genes (TP53, ATR, FANCD2, BAX and BRIP1) and here we discuss how they relate to longevity. The gene exhibiting most interactions in these pathways is TP53 and it seems to act as a driver gene in longevity of Bulgarian centenarians.
TP53 (rs1042522) is a variant designated to be significantly associated with longevity that our results confirm is the case in Bulgarian centenarians. The 2 alleles at the rs1042522 locus encode protein isomorphs that differ in their capacities to induce target gene transcription, their ability to interact with p73 (another tumor suppressor protein) and their targeting of the proteasome. A Danish study finds that minor allele homozygotes, i.e. rs1042522 (C/C) genotypes, live on average 3 years longer than major allele (G/G) homozygotes [15]. The frequency of the C allele in the Bulgarian population is 0.724, similar to European exome frequency 0.738 [16], while the frequency in Bulgarian centenarians is significantly higher (0.844) (see Table 1). The increased longevity is speculated to be related to the increased apoptosis seen for this gain of function SNP [17]. TP53 encodes a tumor suppressor protein which responds to diverse cellular stresses by regulating the expression of genes involved in i.e. inducing cell cycle arrest, DNA repair, or changes in metabolism [18]. Our pathway analysis demonstrated that this gene is set to be involved in several pathways associated with longevity, e.g. regulation of TP53 activity, DNA repair and G2/M DNA damage checkpoint. TP53 directly stimulates transcription of several genes involved in DNA mismatch repair [19-21], in nucleotide excision repair [22], as well as in repairing DNA interstrand crosslinks [23]. Expression of several DNA repair genes is under indirect TP53 control, e.g. genes involved in the repair of DNA double strand breaks and/or the Fanconi anemia pathway [24-27]. Throughout the cell cycle, the genome is constantly monitored for damage, resulting either from errors of, by-products of metabolism or through extrinsic sources such as ultra-violet or ionizing radiation. The different DNA damage checkpoints act to inhibit or maintain the inhibition of the relevant cyclin-dependent kinase that will control the next cell cycle transition.
Our results show significant association between rs1802904 in the ATR gene and longevity, whereas Longevity Map doesn’t. The ATR protein is a serine/threonine kinase which phosphorylates TP53 in stress cells and participates in the post-translational modification of TP53 activity [28]. The ATR gene was found to be involved in the regulation of DNA repair and G2/M DNA damage checkpoint pathways [29], and functions as an apoptotic activator [30]. Failure of the G2 DNA damage checkpoint leads to catastrophic attempts to segregate unrepaired chromosomes. The variant rs1802904 is however synonymous and with unknown functional significance, and further research is needed to clarify its effect on longevity.
The variant rs3172417 in FANCD2 gene is not designated as significantly associated with longevity by Longevity Map, nevertheless our results show significant association. The FANCD2 gene is also involved in the regulation of TP53 activity and DNA repair pathways [31]. Part of the Fanconi anemia complementation group, the protein encoded by this gene is monoubiquinated in response to DNA damage, resulting in its localization to nuclear foci with other proteins (BRCA1 and BRCA2) involved in homology-directed DNA repair [32]. The rs3172417 SNP is in a 3’ untranslated region and little is known about its functional mechanisms, and its association with longevity should be confirmed in a larger case-control studies.
We find significant association with longevity of rs1805419 in the BAX gene, in line with Erdman, Nasibullin [33] who have found that the G allele of this variant is significantly associated with longevity. BAX is a transcriptional target for the TP53 and plays a role in mitochondrial apoptosis. The protein encoded by this gene forms a heterodimer with BCL2 (Bcl2-Bax) with anti-apoptotic function, while Bax/Bax homodimer acts as apoptotic inducer. The factors that provide the fine balance between programmed cell death and cell survival processes probably regulate not only cell longevity but could also be important for human longevity.
Another variant with uncertain association in Longevity Map is rs4986764 in the BRIP1 gene, yet our results indicate significant association. BRIP1 is also involved in regulation of TP53 activity, DNA repair and G2/M DNA damage checkpoint pathways. The protein encoded is a component of a complex important in the normal double-strand break repair function of breast cancer (BRCA1) [34]. Meta-analysis indicates that rs6504074 may play a decreased risk of gynecologic cancer in the overall population [35].
The EGFR variant rs2072454 has previously been associated with longevity in Koreans [36], and we corroborate this significant association. Interaction the EGRF protein with ligands triggers signaling pathways within the cell and leads to DNA synthesis and cell proliferation. Pathogenic mutations in this gene are associated with lung cancer with head and neck squamous cell carcinoma (HNSCC) [37].
IL10 (rs1518111) is an intronic non-coding variant. Its functional consequence on the processing of the transcript and the protein is unknown. The protein produced by this gene is an anti-inflammatory cytokine that has pleiotropic effects in immunoregulation and inflammation. Other IL10 gene polymorphisms has previously been linked to longevity, e.g. the high IL10-producer genotype is increased among centenarians [38].
Pathway analysis of gene set with negative LAVs in Bulgarian population
Unlike the set of genes with positive LAVs which were assigned to be part of gene expression/transcription (GET) network, ten of 20 significant pathways of genes carrying putatively disadvantageous variants for longevity are part of a different significantly over-represented network - the signal transduction (ST) network. The ST network includes the following pathways: Signaling by VEGF resulting angiogenesis by mediating endothelial cell proliferation and vascular permeability; Signaling by receptor tyrosine kinases; G-protein beta:gamma signaling; Signaling by AKT1; Signaling to ERKs; Non-genomic estrogen signaling. These pathways include a combination of 6 genes from our input set: AKT1, HRAS, NOS3, PDPK1, PRKCB and NTRK.
All cells are sensitive to specific signals and can respond to changes in their environment [39]. Signal transduction is the transmission of chemical or physical signals from the cell membrane to the nucleus and coordinates the communication among cells and between cells and extracellular matrix. Finetuned positive and negative regulation of signaling networks is a critical feature of normal, physiological signaling balance in any given cell. Despite the presence of cellular protective responses, pathologies (e.g. cancers) frequently develop in tissues due to somatic mutations within these key signal transduction networks [40,41]. Dysregulation of signal transduction could lead cells to overproliferating and to bypassing survival and migration mechanisms, thus promoting cancer development. During the aging process, changes in the signaling intensities of these networks could manifest in agerelated pathologies [41].
Vascular endothelial growth factor (VEGF) and its receptors (VEGFR) are key regulators of the process of angiogenesis. VEGF/VEGFR signaling pathways include five genes from the set input in REACTOME with negative LAVs in Bulgarian centenarians (AKT1, HRAS, NOS3, PDPK1, and PRKCB). Disturbances in these pathways play role in disease pathology and may be an early step in the process of metastasis, which makes them attractive therapeutic targets.
Signaling by receptor tyrosine kinases is a pathway in which function the same set of 5 genes. Receptor tyrosine kinases (RTKs) are a major class of cell surface proteins involved in common signaling pathways including RAF/MAP kinase cascades [42] and AKT signaling [43]. RTKs regulate many key cell processes and their dysregulation is established in a wide range of cancers. Therefore, RTKs have become attractive therapeutic targets.
G-proteins are involved in transmitting signals from outside the cell to the nucleus through connected signaling cascades, and changes in G proteins and their downstream signaling could initiate cancer. AKT1 is a known oncogene, recognized as a critical node in the PI3K/AKT/mTOR pathway. Reduced activity of this nutrient-sensing pathway is also one of the few physiological mechanisms repeatedly shown to be associated with longevity in humans [44].
The extracellular signal-regulated kinase (ERK) signaling pathway plays role in controlling diverse cellular processes such as proliferation, survival, differentiation and motility. The ERK pathway is often up-regulated in human tumors and as such represents a suitable target for the development of anticancer drugs.
Non-genomic estrogen signaling is well characterized in multiple carcinomas. Recent studies have established a potential immune regulatory role of estrogens in the tumor microenvironment, and promote estrogen as a potential mediator of tumor immunosuppression [45]. Estrogen signaling regulates of gene expression of certain chromatinmodifying enzymes and miRNAs and is connected to epigenetic mechanisms.
Other significant pathways indicated by the set of genes showing prevalence of allele frequency in the control pool constitute the DNA repair and immune system networks, both of which have been shown to impact longevity [46,47].
Variants in the APOE gene
Among variants that showed significant difference in allele frequencies between the centenarians and controls were absent variants in the APOE gene. As this gene has been repeatedly associated with longevity, we consider separately the variants in APOE. The risk C allele in rs429358 was only detected in the control pool and with lower frequency compared to other populations (Table 5).
| Pool | Ref | Alt | Function | dbSNP | Ref. allele (n) | Alt. allele (n) | Frequency alt. allele | Other population source | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Bulgarian | Other populations | |||||||||
| Centenarians | C | G | Exonic | rs440446 | 32 | 50 | 0.61 | 0.641 | gnomAD_genome_NFE | |
| Controls | C | G | Exonic | rs440446 | 31 | 38 | 0.551 | 0.641 | gnomAD_genome_NFE | |
| Centenarians | G | A | Intronic | rs769449 | 737 | 25 | 0.033 | 0.115 | gnomAD_genome_NFE | |
| Controls | G | A | Intronic | rs769449 | 534 | 62 | 0.104 | 0.1145 | gnomAD_genome_NFE | |
| Centenarians | C | T | Intronic | rs143063029 | 903 | 13 | 0.014 | 0.0012 | gnomAD_genome_NFE | |
| Centenarians | C | T | Exonic | rs121918393 | 90 | 2 | 0.022 | 0.0001 | ExAC | |
| Centenarians | G | A | Exonic | rs756564996 | 90 | 2 | 0.022 | 6.49E-05 | gnomAD_genome_ALL | |
| Centenarians | G | A | Exonic | rs767339630 | 132 | 2 | 0.015 | NA | gnomAD_genome_NFE | |
| Controls | G | A | Exonic | rs746382742 | 1226 | 4 | 0.003 | 0.00001 | ExAC | |
| Controls | C | T | Exonic | rs752079771 | 282 | 2 | 0.007 | 0. 00002 | gnomAD_exome_NFE | |
| Controls | T | C | Exonic | rs429358 | 84 | 5 | 0.056 | 0.143 | gnomAD_genome_NFE | |
gnomAD_genome_NFE-Genome database with allele frequencies in non-Finnish European population; gnomAD_genome_ALL-Genome database with allele frequencies in all studied populations; gnomAD_exome_NFE - Exome database with allele frequencies in non-Finnish European population; ExAC - exome database of exome aggregation consortium
Table 5: Allele frequency of variants in APOE gene in Bulgarian population.
The absence of this variant in the centenarian pool is in line with previous studies that show this variant is negatively associated with longevity as it is associated with increased risk for cardiovascular disease and Alzheimer disease. Four variants with low frequencies were found only in the centenarian pool: rs143063029, rs121918393, rs756564996, rs767339630. Even though the minor allele frequencies is higher in Bulgarian centenarians compared to other populations, these rare variants are of unknown clinical significance and are not listed in Longevity Map database. More studies are needed to confirm or reject their association with longevity. Three exonic variants in APOE were established only in the control pool: rs746382742, rs752079771, rs429358.
Conclusion
By employing a WES approach we ensure that all gene variants listed in the LongevityMap database are analyzed. The pool-seq approach has proven to give accurate population allele frequency estimates, especially after appropriate filters are used. The downside of this approach is that it is not possible to determine individual genotypes. The positive and negative LAVs participate in different overrepresented pathways. Variants showing significant enrichment in centenarians are in genes that constitute gene expression/transcription (GET) network with leading role of TP53, interplaying with other genes (ATR, FANCD2, BAX, BRIP1). Genes carrying negative LAVs, on the other hand, are members of the signal transduction (ST) network. The involvement of different networks in positive and negative LAVs should be confirmed with additional studies in other populations. The results of this work confirm the importance of studying truly rare survival to discover those combinations of variants associated with extreme longevity and longer health span in genetically different populations. Complex phenotypes such as longevity require not only population level genomic data, but future studies should include GWAS studies of longevous families, functional analysis of interesting loci, somatic mutations, epigenetic studies and microbiomedata.
Acknowledgments
The Bulgarian centenarians project was funded by the National Science Fund of Bulgaria, contract number DN 03/7/18.12.2016
Conflict of Interest
The authors declare that they have no conflict of interest.
Authors Contribution
Dimitar Serbezo, Lubomir Balabanski are equally contributed to the present study.
REFERENCES
- Herskind AM, McGue M, Holm NV, Sorensen TIA, Harvald B, Vaupel JW. The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870-1900. Hum Genet. 1996;97(3): 319-323.
- Garatachea N, Marin PJ, Santos-Lozano A, Sanchis-Gomar F, Emanuele E, Lucia A. The ApoE gene is related with exceptional longevity: a systematic review and meta-analysis. Rejuvenation Res. 2015;18(1): 3-13.
- Dato S, Soerensen M, De Rango F, Rose G, Christensen K, Christiansen L, et al. The genetic component of human longevity: New insights from the analysis of pathway-based SNP-SNP interactions. Aging cell. 2018;17(3): e12755.
- Deelen J, Uh H-W, Monajemi R, van Heemst D, Thijssen PE, Böhringer S, et al. Gene set analysis of GWAS data for human longevity highlights the relevance of the insulin/IGF-1 signaling and telomere maintenance pathways. Age 2013;35(1): 235-249.
- Fracassetti M, Griffin PC, Willi Y. Validation of Pooled Whole-Genome Re-Sequencing in Arabidopsis lyrata. PloS one. 2015;10(10): e0140462.
- Barrio AM, Lamichhaney S, Fan G, Rafati N, Pettersson M, Zhang H, et al. The genetic basis for ecological adaptation of the Atlantic herring revealed by genome sequencing. eLife. 2016;5: e12081.
- Schlotterer C, Tobler R, Kofler R, Nolte V. Sequencing pools of individuals - mining genome-wide polymorphism data without big funding. Nat Rev Genet. 2014;15(11): 749-763.
- Budovsky A, Craig T, Wang J, Tacutu R, Csordas A, Lourenco J, et al. LongevityMap: a database of human genetic variants associated with longevity. Trends Genet. 2013;29(10): 559-560.
- Yang H, Wang K Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc. 2015;10: 1556-1566.
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv 2019: 531210.
- Benjamini Y, Hochberg Y Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Series B Stat Methodol. 1995;57(1): 289-300.
- Team RC. R: A language and environment for statistical computing. 2018;Available from: https://www.R-project.org/.
- Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018; 46(D1): D649-D655.
- Fang X, Sastry A, Mih N, Kim D, Tan J, Yurkovich JT, et al. Global transcriptional regulatory network for Escherichia coli robustly connects gene expression to transcription factor activities. Proc Natl Acad Sci U S A. 2017;114(38):10286-10291.
- Bojesen SE, Nordestgaard BG. The common germline Arg72Pro polymorphism of p53 and increased longevity in humans. Cell cycle.2008;7(2): 158-163.
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature.2016;536:285-291.
- Huang X, Wu F, Zhang Z, Shao Z Association between TP53 rs1042522 gene polymorphism and the risk of malignant bone tumors: a meta-analysis. Biosci Rep.2019;39(3): BSR20181832.
- Hafner A, Bulyk ML, Jambhekar A, Lahav G. The multiple mechanisms that regulate p53 activity and cell fate. Nat Rev Mol Cell Biol.2019;20(4): 199-210.
- Scherer SJ, Maier SM, Seifert M, Hanselmann RG, Zang KD, Muller-Hermelink HK, et al. p53 and c-Jun functionally synergize in the regulation of the DNA repair gene hMSH2 in response to UV. J Biol Chem.2000; 275(48): 37469-37473
- Warnick CT, Dabbas B, Ford CD, Strait KA. Identification of a p53 response element in the promoter region of the hMSH2 gene required for expression in A2780 ovarian cancer cells. J Biol Chem. 2001;276(29): 27363-27370.
- Chen J, Sadowski I. Identification of the mismatch repair genes PMS2 and MLH1 as p53 target genes by using serial analysis of binding elements. Proc Natl Acad Sci U S A. 2005;102(13): 4813-4818.
- Tan T, Chu G. p53 Binds and activates the xeroderma pigmentosum DDB2 gene in humans but not mice. Mol cell Biol.2002; 22(10):3247-3254.
- Liebetrau W, Budde A, Savoia A, Grummt F, Hoehn H. P53 activates Fanconi anemia group C gene expression. Human Mol Genet 1997;6(2): 277-283.
- Blazek D, Kohoutek J, Bartholomeeusen K, Johansen E, Hulinkova P, Luo Z, et al. The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes & Dev. 2011;25(20): 2158-2172.
- Zhe C, Wei W,Yong-Na S, Yan S,Jie X. hOGG1, p53 genes, and smoking interactions are associated with the development of lung cancer. Asian Pac J Cancer Prev. 2012;13(5): 1803-1808.
- Ekumi KM, Paculova H, Lenasi T, Pospichalova V, Bösken CA, Rybarikova J, et al. Ovarian carcinoma CDK12 mutations misregulate expression of DNA repair genes via deficient formation and function of the Cdk12/CycK complex. Nucleic acids Res. 2015;43(5): 2575-2589.
- Bartkowiak B, Greenleaf AL. Expression, purification, and identification of associated proteins of the full-length hCDK12/CyclinK complex. J Biol Chem. 2015;290(3): 1786-1795.
- Maréchal A, Zou L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol. 2013;5(9): a012716.
- Rocha S, Garrett MD, Campbell KJ, Schumm K, Perkins ND. Regulation of NF‐κB and p53 through activation of ATR and Chk1 by the ARF tumour suppressor.EMBO J. 2005;24(6):1157-1169.
- Minton K. ATR prevents premature apoptosis. Nat Rev Mol Cell Biol. 2015;16: 640-641.
- Jaber S, Toufektchan E, Lejour V, Bardot B, Toledo F. p53 downregulates the Fanconi anaemia DNA repair pathway. Nat Commun. 2016;7: 11091.
- Kennedy RD, D'Andrea AD. The Fanconi Anemia/BRCA pathway: new faces in the crowd. Genes & Dev. 2005;19(24): 2925-2940.
- Erdman VV, Nasibullin TR, Tuktarova IA, Mustafina OE. Association of polymorphic markers of CASP8, BCL2, and BAX genes with aging and longevity. Adv Geronto. 2013;3(2): 93-99.
- Zona S, Bella L, Burton MJ,De Moraes GN, Lam EWF. FOXM1: an emerging master regulator of DNA damage response and genotoxic agent resistance. Biochim Biophys Acta Gene Regul Mech. 2014;1839(11): 1316-1322.
- Sigurdson AJ, Hauptmann M, Chatterjee N, Alexander BH, Doody MM, Rutter JL, et al (2004) Kin-cohort estimates for familial breast cancer risk in relation to variants in DNA base excision repair, BRCA1 interacting and growth factor genes. BMC cancer. 2004;4: 9.
- Park JW, Ji YI, Choi YH, Kang MY, Jung E, Cho SY, et al. Candidate gene polymorphisms for diabetes mellitus, cardiovascular disease and cancer are associated with longevity in Koreans. Exp Mol Med. 2009;41(11): 772-781.
- Fung C, Zhou P, Joyce S, Trent K, Yuan JM, Grandis JR, et al. Identification of epidermal growth factor receptor (EGFR) genetic variants that modify risk for head and neck squamous cell carcinoma. Cancer Lett. 2015;357(2): 549-556.
- Caruso C, Lio D, Cavallone L, Franceschi C. Aging, longevity, inflammation, and cancer. Ann N Y Acad Sci. 2006;1028(1): 1-13.
- Duan G, Walther D. The roles of post-translational modifications in the context of protein interaction networks. PLoS Comput Biol. 2015;11(2):e1004049.
- Moon RT, Kohn AD, De Ferrari GV, Kaykas A. WNT and beta-catenin signalling: diseases and therapies. Nat Rev Genet. 2004;5(9): 691-701.
- Katoh M. Networking of WNT, FGF, Notch, BMP, and Hedgehog signaling pathways during carcinogenesis. Stem cell Rev. 2007;3(1): 30-38.
- McKay MM, Morrison DK. Integrating signals from RTKs to ERK/MAPK. Oncogene 2007;26(22): 3113-3121.
- Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell 2007;129(7): 1261-1274.
- Ostan R, Monti D, Gueresi P, Bussolotto M, Franceschi C, Baggio G. Gender, aging and longevity in humans: an update of an intriguing/neglected scenario paving the way to a gender-specific medicine. Clinical science. 2016; 130(19): 1711-1725.
- Rothenberger NJ, Somasundaram A, Stabile LP. The Role of the Estrogen Pathway in the Tumor Microenvironment. Int J Mol Sci. 2018; 19(2): 611.
- Maluf SW, Martínez-López W, Da Silva J. DNA Damage: Health and Longevity. Oxid Med Cell Longev. 2018; 9701647.
- Patricia A, Monica DF. Role of the immune system in aging and longevity. Curr Aging Sci. 2011;4(2): 78-100.
Citation: Serbezov D, Balabanski L, Karachanak-Yankova S, Vazharova R, Nesheva D, Hammoudeh Z, et al. (2019) POOL-SEQ Study of Bulgarian Centenarians Highlights the relevance for Human Longevity of Gene Expression Pathways. J Aging Sci 7: 2. doi: 10.35248/2329-8847.19.7.208
Copyright: © 2019 Serbezov D, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.