PMC/PubMed Indexed Articles
Indexed In
- Academic Journals Database
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- ResearchBible
- Ulrich's Periodicals Directory
- Electronic Journals Library
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- MIAR
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2019) Volume 10, Issue 1
Nitrogen Base Sequence Analysis and Characterization of Mutations in Gene Coding Region That Can Lead to High Levels of Resistance in Tuberculosis Patients in Jayapura, Papua Province-Indonesia
Rosye H. R. Tanjung1 and Yohanis Ngili2*2Faculty of Mathematics and Natural Sciences, Department of Chemistry, Biochemistry Research Group, Cenderawasih University, Jayapura, Indonesia
Received: 28-Jan-2019 Published: 25-Mar-2019
Abstract
The disease of tuberculosis in Papua Province, Indonesia, is very high based on data at the Papuan Provincial Health Department. The geographical and demographic conditions of the Papuan population cause the eradication of the disease more difficult. One cause of the development of this disease is the presence of anti-tuberculosis drug resistance. This study was conducted by analyzing samples from sputum of TB patients. The increasing number of HIV/AIDS sufferers has caused TB disease, WHO categorize as reemerging disease, especially in Papuan province of Indonesia, the number of people with HIV/AIDS is highest in Indonesia. The purpose of this research is to get information on MDR-TB relationship with coding genes, and review the results of several studies of M. tuberculosis genotype on isolates in Jayapura, Papua Province-Indonesia. Here, we reported that a change in nucleotide C1363A (Pro535His) in sensitive M. tuberculosis of several antituberculosis drugs demonstrated that only a few mutations in the rpob gene caused resistant properties. The results of this research open a new path paradigm focused on mutations in the area of gene promoter and noncoding region.
Keywords
Characterization; Rpob512; RNA polymerase β-subunit resistance; Tuberculosis; Papua-Indonesia Province
Introduction
The provincial government of Papua, Indonesia, has passed various methods and techniques to reduce the high number of tuberculosis diseases. Various programs are conducted to inhibit lanju transmission of this disease and even suppress the number of drug resistance in patients. TB disease is characterized by tissue death (necrosis) caused by slow type hypersensitivity, namely the phagocytic process and epitope presentation (identification of antigens) by macrophage cells on the surface of the cell resulting in a series of processes that trigger the reaction of T lymphocyte cells. continues to increase in TB treatment and control is a multidrug-resistant M. tuberculosis (MDR-TB) isolate, defined by the world health agency WHO as a RIF and INHresistant M. tuberculosis isolate. Treatment of TB patients is usually done by administering three types of antituberculous drugs with the main options being rifampin (RIF) and isoniazid (INH), then accompanied by streptomycin or pyrazinamide [1].
As the number of HIV/AIDS sufferers increases, WHO categorizes TB disease as reemerging disease. As a result of rpoB mutations, especially in hotspots or RRDR (rifampin resistancedetermining region), RIF cannot inhibit RNA polymerase because it cannot bind to the β-subunit. Meanwhile, INH requires an activation process by the catalase-peroxidase enzyme that produced by M. tuberculosis [2,3]. Data from the provincial health offices of Papua-Indonesia, showing more than 95% resistant of M. tuberculosis-caused by mutation of rpoB and approximately 65% of M. tuberculosis resistant to INH are caused by katG mutations. This study was conducted by analyzing samples from sputum TB patients in Papua Province-Indonesia [4].
Materials and Methods
Isolation, characterization of phenotype and genotype characterization
Samples were obtained from Dok 2 Jayapura hospital and Abepura General Hospital in Jayapura-Papua-Indonesia Province. Suspension isolates are made by introducing M. tuberculosis colonies into sterile tubes containing physiological and glassbead solutions so as to achieve turbidity of McFarland. This process is done in Biochemistry Laboratory, Department of Chemistry, University of Cenderawasih, Jayapura-Papua, Indonesia. Genotypic characterization process was carried out to analyze 4 genes of M. tuberculosis, two genes that produced membrane proteins and the other two were rpoB gene and other secondary areas, such as katG which caused the resistance of M. tuberculosis to RIF and INH. The result of this primer pair amplification is called the non-variable band because it must always exist to mark the running of the multiplex PCR process [5-8].
Determining the sequence of other gene nucleotides is done to see how these genes relate to MDR-TB resistant traits. Research on the coding region of this gene performed on several MDR-TB isolates in Papua province represented high-grade resistance and low resistance levels. Determination of this nucleotide sequence has been done by using the services of Macrogen Inc., Seoul, South Korea. The entire sequence of nucleotide sequencing data was analyzed by DNAstar program using various applications in it as well as some applications used in Biochemistry Research Group, University of Cenderawasih.
Results and Discussion
Analysis of rpoB genotype
The rpoB gene has a length of 4.8 kb while the rpoB gene nucleotide sequence analysis is performed successively on the nucleotide sequence 1.5-1.7 kb. It is possible that the amount of printed DNA added to the PCR mixture is too much so that the PCR reaction shows non-specific results. The use of three primers on multiplex PCR causes the temperature for primer adhesion to be more than one type. The presence of non-specific bands on PCR multiplex results did not affect genotype analysis for rpoB531, and rpoB526 because this technique was only used to study the existence of a variable DNA fragment band of 0.18 kb and 0.17 kb for the codon: rpoB526 and rpoB531. DNA samples used in multiplex PCRs are the result of cell lysis that is not measured by its DNA concentration, causing the DNA concentrations in each sample of multiplex PCR reactions to vary. Approximately more than 90% of RIF-resistant isolates are caused by the mutation of rpoB gene in the region of 81 pb. This area is called a hotspot area and known as RRDR [9-12]. The frequent mutations caused changes i.e., (TCG become TTG) and (CAC become GAC) or Ser531Leu and His526Asp [13].
The results of this study indicate that approximately 85 MDRTB isolates are known to have multiplexed PCR mutations, while 19% of undetectable MDR-TB isolates undergo rpoB526 or rpoB531 mutations. Different results were obtained from P3 isolate samples and some isolates from several hospitals in Jayapura, i.e., no mutation with multiplex PCR rpoB526 but electroferogram result showed adenin mutation to guanine at position 1576, A1576G (rpoB526 codon).
A number of 12 amino acid residues in RNA polymerase β- subunit are involved in interaction with RIF. The substitution of 11 out of the 12 amino acid residues will give rise to RIFresistant properties [14-16] mentioning that more than 90% of RIF resistance occurs due to a genetic change in the 81 bp RRDR fragment of rpoB gene encoding the β-subunit RNA polymerase. Meanwhile, several studies have mentioned mutations of rpoB gene outside the RRDR can also cause resistance.
G1389C mutated mutations occur in isolates P5, P11, and P12 so it is suspected that this mutation is a form of rpoB M. tuberculosis gene polymorphism in addition to A1538T on isolate P1. Other bioinformatics researchers suggests that changes in rpoB gene residues at at RRDR 511, 512, 515, 521, and 529 positions do not significantly affect RIF's minimum inhibitory concentration MIC, but this study suggests that rpoB512 mutations can also cause high levels of resistance. Meanwhile, other research results suggest a change in 12 amino acids in the region (pocket) proteins resulted in the region being not in direct contact with the RIF.
The analysis of the determination of nucleotide sequence of isolate P4 was done in two directions, using primary and back primers and both showed the same result. Other studies were also conducted on the same genes [rpoB516]. The resistive nature of the RIF of the M. tuberculosis isolate acquired by Tracevska et al., is the probable outcome of [rpoB516], not [rpoB535] that the result of Tracevska et al., may be used to support the possibility [rpoB535] - which does not give rise to M. tuberculosis (Figure 1) [17-19].
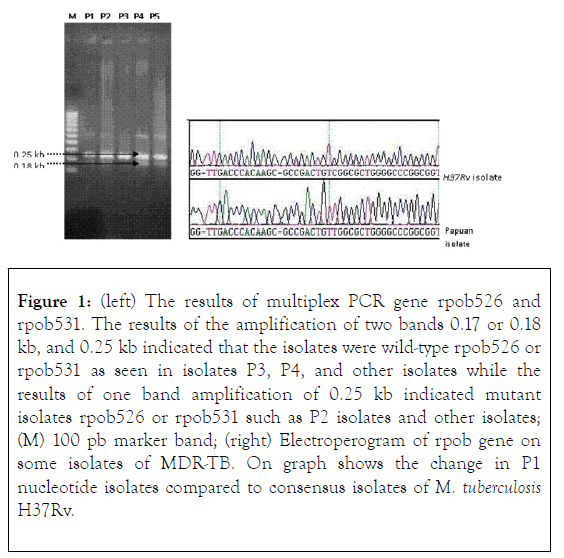
Figure 1. (left) The results of multiplex PCR gene rpob526 and rpob531. The results of the amplification of two bands 0.17 or 0.18 kb, and 0.25 kb indicated that the isolates were wild-type rpob526 or rpob531 as seen in isolates P3, P4, and other isolates while the results of one band amplification of 0.25 kb indicated mutant isolates rpob526 or rpob531 such as P2 isolates and other isolates; (M) 100 pb marker band; (right) Electroperogram of rpob gene on some isolates of MDR-TB. On graph shows the change in P1 nucleotide isolates compared to consensus isolates of M. tuberculosis H37Rv.
Changes in rpoB526 and rpoB531 are mutations in the RRDR region which are widely known because they are most dominant in RIF-resistant isolates. This study obtained 16 (38.1%) and 15 isolates (35.7%) for [rpoB526]- and [rpoB531]-, 3 isolates (7.1%) double mutants [rpoB526, rpo531]-, 3 isolates experienced mutations in other rpoB codons, and 5 isolates not detected experienced mutations in the rpoB segment analyzed, out of 42 MDR-TB isolates analyzed. The results of the Cummings et al., (2004) study of 103 RIF-resistant isolates showed 53, 31, and 20 isolates respectively for [rpoB531]- [rpoB526]- and other rpoB mutants. Meanwhile, only the katG315 mutation is known to cause resistance to INH, and this study only received 16 isolates [katG315]-.
The results of genotype characterization in the rpoB gene carried out in this study were lower than those reported by Pozzi et al., (1999) who conducted research in Italy, namely 56.7% of the rpoB531 mutation and 24.3% of the rpoB526 mutation occurred in isolates MDR-TB. Yue et al., (2003) who conducted research in China obtained 41% of MDR-TB isolates were [rpoB531]-, and the other 40% were [rpoB526]-. The research of Yue et al. is not different from the results of this study, namely the frequency [rpoB526]- and [rpoB531]- which occurs relatively balanced.
The primers we use in this study are as follows in (Table 1).
| Primer Name | Nucleotide Sequence | Temperature (°C) |
|---|---|---|
| Forward primer rpoB | 5’-GTCGCCGCGATCAAGGA-3’ | 56 |
| Reverse primer rpoB | 5’-TGACCCGCGCGTACAC-3’ | 56 |
| Inner primer rpoB531 | 5’-ACAAGCGCCGACTGT C-3’ | 48 |
| Inner primer rpoB526 | 5’-GTCGGGGTTGACCCA-3’ | 50 |
| Forward primer katG | 5’-GCAGATGGGGCTGATCTACG-3’ | 64 |
| Reverse primer katG | 5’-AACGGGTCCGGGATGGTG-3’ | 60 |
| Inner primer katG315 | 5’-ATACGACCT CGATGCCGC-3’ | 62 |
Table 1: Primer nucleotide sequences used in multiplex PCR.
Conclusion
This study suggests that rpoB512 mutations can also cause high levels of resistance. Meanwhile, a change in some of the amino acids in the pocket area of the protein causes the area to be out of direct contact with the RIF. Most of the mutations occurring in the β-subunit RNA polymerase are found in region I (the position of amino acid residues 505 to 537) and region II (the position of amino acids 562 to 572). The results of the structural model analysis of RNA polymerase β-subunit proteins that bind RIF to T. aquaticus, show only a few amino acid residues that bind directly to RIF because they have the same polarity and can form bonds between nitrogen or oxygen, with RIF hydroxyl groups. Amino acid changes in this residue caused the greatest effect on the M. tuberculosis phenotype on the Mtb isolate in Papua Province of Indonesia.
Acknowledgments
The authors gratefully acknowledge the support of this work by the Head and colleagues of Health Laboratory of Jayapura, Papua province of Indonesia, Chairperson of the Laboratory of Biochemistry and Molecular Biology, Faculty of Mathematics and Natural Sciences, University of Cenderawasih that has provided research facilities.
REFERENCES
- Glynn JR, Whiteley J, Bifani PJ, Kremer K, Dick Van Solingen. Worldwide Occurrence of Beijing/W Strains of Mycobacterium tuberculosis: A Systematic Review. Emerg Infect Dis. 2002;8:843-850.
- Mikhailovich V, Lapa S, Gryadunov D, Sobolev A, Strizhkov B, Chernyh N, et al. Identification of Rifampin-Resistant Mycobacterium tuberculosis Strains by Hybridization, PCR, and Ligase Detection Reaction on Oligonucleotide Microchips. J Clin Microbiol. 2001;39:2531-2540.
- Pierattelli R, Banci L, Eady NAJ, Bodiguel J, Jones JN, Moody PCE, et al. Crystal structure of Mycobacterium tuberculosis catalaseperoxidase. J Biol Chem. 2004;279: 39000-39009.
- Ramaswamy SV, Dou SJ, Rendon A, Yang Z, Cave MD, Graviss EA. Genotypic analysis of multidrug-resistant Mycobacterium tuberculosis isolates from Monterrey, Mexico. J Med Microbiol. 2004;53:107-113.
- Ubyaan R, Maryuni AE, Sroyer A, Palit EIY, Jukwati, Suaka IY, et al. Molecular Analysis of Rifampin-Resistant Mycobacterium tuberculosis Strains Isolated from Papua, Indonesia. Int J PharmTech Res 2012;4(4):1803-1811.
- Mokrousov I, Otten T, Filipenko M, Vyazovaya A, Chrapov E, Limeschenko E, et al. Detection of Isoniazid-Resistant Mycobacterium tuberculosis Strains by a Multiplex Allele-Specific PCR Assay Targeting katG Codon 315 Variation. J Clin Microbiol. 2002; 40:2509-2512.
- Korzheva N, Mustaev A, Kozlov M, Malhotra A, Nikiforov V, Goldfarb A, et al. A Structural Model of Transcription Elongation. Science. 2000;289:619-625.
- Putman M, van Veen HW, Konings WN. Multidrug resistance mediated by a bacterial homolog of the human multidrug transporter MDR1. Proc Natl Acad Sci. 1996;93:10668-10672.
- Van der Zanden AGM, Te-Koppele Vije EM, Bhanu NV, Van Soolingen D, Schouls LM. Multidrug resistance mediated by a bacterial homolog of the human multidrug transporter MDR1. J Clin Microbiol. 2003;41:1101-1108.
- Yue J, Shi W, Xie J, Li Y, Zeng E, Wang H. Mutations in the rpoB Gene of Multidrug-Resistant Mycobacterium tuberculosis Isolates from China. J Clin Microbiol. 2003;41:2209-2212.
- Corbett EL, Watt CJ, Walker N, Maher D, Williams BG, Raviglione MC, et al. The growing burden of tuberculosis: global trends and interactions with the HIV epidemic. Arch Intern Med. 2003;163:1009-1021.
- Tanjung RHR and Ngili Y. Exploration of the components of ethanol extract of herbal medicine origin of Papua Province-Indonesia that interact with the DNA of Escherichia coli. Der Pharma Chemica. 2016;8(6):165-173.
- Hirano K, Abe C, Takahashi M. Mutations in the rpoB Gene of Rifampin-Resistant Mycobacterium tuberculosis Strains Isolated Mostly in Asian Countries and Their Rapid Detection by Line Probe Assay. J Clin Microbiol. 1999;37: 2663-2666.
- Tanjung RHR and Ngili Y. Nucleotide Sequences and Mutations in Katg Gene in Clinical Isolates of Mycobacterium tuberculosis Isolates Resistant to Isoniazid in Papua-Indonesia. Int J PharmTech Res. 2016;9(5):334-341.
- Höfling CC, Pavan EM, Giampaglia CMS, Ferrazoli L, Aily DCG, de Albuquerque DM. Prevalence of katG Ser315 substitution and rpoB mutations in isoniazid-resistant Mycobacterium tuberculosis isolates from Brazil. Int J Tuberc Lung Dis. 2005;9(1):87–93.
- Kawulur HSI and Ngili Y. Analysis and Mutation of Codon in rpoB and katG Genes and Bioinformatics Study of RIF Binding Model by RNA β Polymerase Subunit: Study in Tuberculosis Patients at Merauke General HospitalIndonesia. J Data Mining Gen Proteomics. 2018;9(1):1.
- Rie AV, Warren R, Mshanga I, Jordaan AM, Spuy GD, Richardson M, et al. Analysis for a Limited Number of Gene Codons Can Predict Drug Resistance of Mycobacterium tuberculosis in a High-Incidence Community. J Clin Microbiol. 2001;39:636-641.
- Wei CJ, Lei B, Musser JM, Tu SC. Isoniazid Activation Defects in Recombinant Mycobacterium tuberculosis Catalase-Peroxidase (KatG) Mutants Evident in InhA Inhibitor Production. Antimicrob Agents Chemother. 2003;47:670-675.
- Siallagan J, Maryuni A, Jukwati, Ngili Y. Single nucleotide mutations of intergenic and intragenic region in mitochondrial genome from different individuals from Papua-Indonesia. Der Pharma Chemica. 2015;7(9):334-339.
Citation: Tanjung RHR, Ngili Y (2019) Nitrogen Base Sequence Analysis and Characterization of Mutations in Gene Coding Region That Can Lead to High Levels of Resistance in Tuberculosis Patients in Jayapura, Papua Province-Indonesia. J Data Mining Genomics Proteomics 10:1. doi: 10.4172 /2153-0602.1000217
Copyright: Ã?Æ?Ã?â??Ã?â??Ã?© 2019 Tanjung, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.
Sources of funding : The authors gratefully acknowledge the support of this work by the Head and colleagues of Health Laboratory of Jayapura, Papua province of Indonesia, Chairperson of the Laboratory of Biochemistry and Molecular Biology, Faculty of Mathematics and Natural Sciences, University of Cenderawasih that has provided research facilities.


