PMC/PubMed Indexed Articles
Indexed In
- Academic Keys
- ResearchBible
- CiteFactor
- Access to Global Online Research in Agriculture (AGORA)
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2020) Volume 9, Issue 2
Mutational analysis of FOG2 gene in patients with congenital Heart disease
Brindha Elangovan*Received: 15-Jul-2020 Published: 03-Aug-2020, DOI: 10.35248/2329-6682.20.9.152
Abstract
Congenital heart disease (CHD) is one of the types of developmental defect, with high rates of morbidity in infants. A transcription factor GATA binding factor 4 (GATA4) has been reported to serve an important role in embryogenesis and cardiac development. Aim of the study is to evaluate Mutational analysis of FOG2 genein congenital Heart disease. We have chosen FOG2 for the identification of rare variations in CHD patients from Indian population. To evaluate the study we have done the PCR amplification for Exon 7 and Exon 8, since most of the zinc finger domains of FOG2 are localized in these two exons. Exon 8 was subdivided into 5 different region i.e. Exon 8A-8E because it has long coding region. These exons were amplified in different conditions. For example, Exon 8A-B and E were amplified with Platinum taq DNA polymerase without requirement of any PCR enhancers such as DMSO (Di-methyl-sulfo-oxide) or betaine. However, Exon 8C was amplified in the presence of DMSO. The annealing temperature for every exons are different. Exon 7 is amplified Taq DNA polymerase. The specificity and quantity of amplified products are checked by the 100bp DNA ladder.
Keywords
Cardiovasculardisease-mutation; FOG2; Gene-zinc; Finger protein-Taq; DNA polymerase
Introduction
Congenital heart disease (CHD) remains a leading cause of deathin newborn and infants and is one of the most common human birth defect, affecting nearly 1% of all live births worldwide (Hoffman and Kaplan) [1]. However, several hospital and community based studies have been carried out in last two decades throughout the country to represent the actual burden of the CHD in the society. The etiology of CHD is poorly understood. It may be due to mutations in the genes involved in embryonic heart development during embryogenesis. Chromosomal anomalies including numerical and structural anomalies may also play causative role in the CHD (Pierpont et al) [2]. Although, the knowledge about the role of mutations and chromosomal anomalies in CHD are increasing gradually, the role play by environmental factors cannot be overlooked completely. Prenatal exposure to Angiotensin Converting Enzyme (ACE) inhibitors increases the risk of several congenital defects, including those that cause heart diseases (Cooper et al.) [3]. Generally, congenitalheart diseases arise through various combinations of genetic and environmental factors. The resources that support the contribution of specific environmental factors in CHD causation are limited still there are many examples like folic acid supplementation in the preand peri-conception period, rubella vaccination before pregnancy and proper nutrition may reduce the risk of CHD in infants (Bruneau) [4].
There are many signaling pathways such as BMPs, WNTs/ β- catenin, TGF-β, Notch signaling and Hedgehog signaling, transcription factors such as NKXs, GATAs, TBXs, SRF, FGFs and transcriptional regulators that are involved in the regulation processes. However, the roles of these signaling pathways in the early cardiac development have been well studied, but still it is in its infancy. TGF-β superfamily is a large group of molecules including more than 30 molecules such as TGF-β, NODAL, BMPs, Activins, GDFs, AMH, LEFTY etc. that are involved in the specification, determination of cardiac development. Transforming growth factor β (TGFβ) superfamily is known to play a crucial role in early embyonic development (Rochais et al.) [5].
Mutations in several members of TGFβ superfamily such as NODAL, CRIPTO, ACVR2B and LEFTY has been reported in CHD cases. Besides these, several other transcription factors i.e. NKX2-5, GATA4 and TBX5 and TBX20 etc. are also known as causative genes in the CHD (Schott et al.) [6]. Several hundred genes have been implicated in the formation of the heart, and a mutation in any of them could potentially contribute to a cardiac defect. Identifying which of these genes is involved in human congenital heart disease has been a challenge for scientists in the field. Over the past couple of decades, there has been a greater understanding of the molecular pathways regulating cardiac development.
The development of gene targeting technology has led to the generation of a multitude of mouse models with cardiac developmental defects. Congenital heart disease refers to a range of possible heart defects. The following defects are Aortic Valve Stenosis(AS) and Pulmonary Valve Stenosis (PS), Coarctation of The Aorta (AC),Ebstein's Anomaly, Patent Ductus Arteriosus (PDA), Septal Defects–including atrial septal defects (ASD) and ventricular septal defects (VSD), Single Ventricle Defects – including tricuspid atresia and hypoplastic left heart syndrome (HLHS), Tetralogy Of Fallot (TOF), Total Anomalous Pulmonary Venous Connection (TAPVC), Transposition Of The Great Arteries (TGA), Truncus Arteriosus (TA).
Due to their diversity, it is difficult to come up with a simple definition of what unites all ZnF proteins; however, the most common approach is to define them as all small, functional domains that require coordination by at least one zinc ion (Laity et al.) [7]. Various ZnF motifs will bind DNA, usually in a sequence-specific manner. DNA-binding ZnF proteins are involved in transcriptional processes. Many transcription factors make DNA contacts to gene promoters via ZnFdomains, some of which are classical C2H2ZnF motifs.ZnF motifs typically wrap around the outside of a DNA double helix, following a righthanded helical path.Multiple DNA contacts are made, often involving more than one ZnF domain.These contacts are usually made to bases in the major DNA groove; for example, C2H2ZnFsrecognise DNA sequences by binding to the major groove of DNA via a short alpha-helix in the ZnF, the Znf spanning 3-4 bases of the DNA. There are many DNA-binding proteins with ZnF motifs, several of which are involved in gene transcription.For example, transcription factor IIIA (TFIIIA) is an essential component of the initiation complex that interacts with separate specific transcription signals located within the ribosomal 5S RNA gene promoter.
Non-binding spacers within TFIIIA allow it to span long DNA sequences, binding only to specific regions within the DNA. Several synthetic ZnF proteins have been constructed as novel DNA-binding proteins with specific DNA recognition properties. Transcriptional activator that binds to the consensus sequence 5'-AGATAG-3' and plays a key role in cardiac development and function. In cooperation with TBX5, it binds to cardiac super-enhancers and promotes cardiomyocyte gene expression, while it downregulates endo-cardial and endothelial gene expression. Transcriptional activators are involved in bone morphogenetic protein (BMP)-mediated induction of cardiacspecific gene expression Figures 1 and 2.
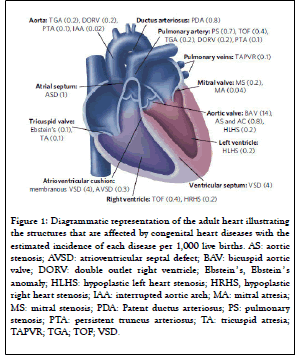
Figure 1: Diagrammatic representation of the adult heart illustrating the structures that are affected by congenital heart diseases with the estimated incidence of each disease per 1,000 live births. AS: aortic stenosis; AVSD: atrioventricular septal defect; BAV: bicuspid aortic valve; DORV: double outlet right ventricle; Ebstein ’ s, Ebstein ’ s anomaly; HLHS: hypoplastic left heart stenosis; HRHS, hypoplastic right heart stenosis; IAA: interrupted aortic arch; MA: mitral atresia; MS: mitral stenosis; PDA: Patent ductus arteriosus; PS: pulmonary stenosis; PTA: persistent truncus arteriosus; TA: tricuspid atresia; TAPVR; TGA; TOF; VSD.
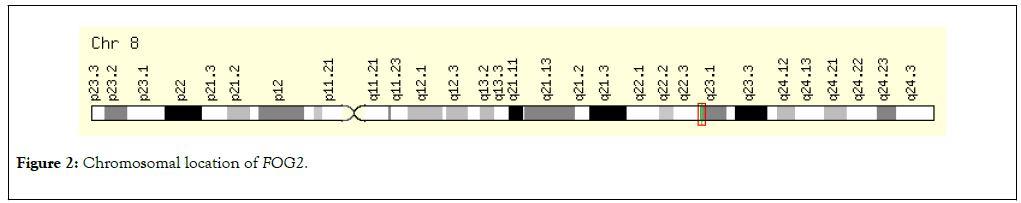
Figure 2: Chromosomal location of FOG2. Figure
BMP response elements (BMPRE) are involved in DNA sequences within cardiac activating regions (by sequence similarity). These acts as a transcriptional activator of ANF in cooperation with NKX2-5 (by similarity), promotes cardiac myocyte enlargement. GATA4 is a member of the GATA family of zinc finger transcription factor, which regulates gene transcription by binding to GATA elements and plays a central role in cardiac development and is critical for survival of the embryo (Peterkin et al.) [8].
Dosage of GATA4 is responsible for regulation of cardiac morphogenesis, and graded reduction of GATA4 leads to abnormal cardiac development with a common AVC, double outlet right ventricle, and hypoplasia of the ventricular myocardium (Pu et al.) [9]. GATA4 is also an important regulator of cardiomyocyte proliferation through direct transcriptional activation of cell cycle regulators, including cyclin D2 and cdk4, which may explain the lack of sufficient cardiac septation and chamber hypoplasia observed in patients with GATA4 mutations(Rojas et al.) [10]. The zinc finger protein encoded by this gene is a widely expressed member of the FOG family of transcription factors. The family members modulate the activity of GATA family proteins, which are important regulators of hematopoiesis and cardiogenesis in mammals. It has been demonstrated that the protein can both activate and down-regulate expression of GATA-target genes, suggesting different modulation in different promoter contexts Figures 3 and 4.

Figure 3: Diagramatic representation of interaction between FOG2 and GATA.
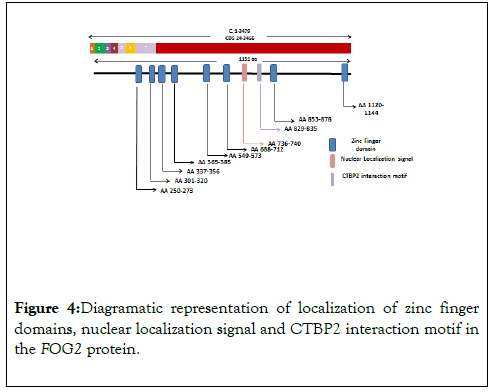
Figure 4: Diagramatic representation of localization of zinc finger domains, nuclear localization signal and CTBP2 interaction motif in the FOG2 protein.
The zinc finger protein encoded by FOG2 gene is a widely expressed member of the FOG family of transcription factors. It consists of total eight exons which encode an 1151 amino acid long nuclear protein that contains eight zinc finger domains. These zinc finger domains are concentrated in last two exons i.e. two zinc finger domains are present in exon 7 while six domains are located in the exon 8. Distribution of Zinc finger and other interacting domains are diagrammatically presented in Figure 1. FOG2 interacts directly with the N-terminal zinc finger of GATA4 and plays a central role in heart morphogenesis by regulating genes that are essential during cardiogenesis (Nikolay et al.) [11].
Materials and Methods
Polymerase chain reaction (PCR)
Genomic DNA isolated from blood sample of congenital heart disease patients were amplified using polymerase chain reaction. Polymerase chain reaction is used to amplify a single or a few copies of a piece of DNA across several orders of magnitude, generating thousands to millions of copies of particular DNA sequence.
Reagents for Polymerase chain reaction: Polymerase chain reaction requires DNA template, oligonucleotide primers, Taq Polymerase, PCR buffer, deoxy-nucleotide tri-phosphates (dNTPs), MgCl2 and de-ionized water. The working concentration of each constituent Tables1 and 2.
| S.No. | Reagent | Stock Concn | Working Concn | Amount (µl)/reaction |
|---|---|---|---|---|
| 1. | DNA Template | 10 ng/ µl | 1ng/ µl | 2 |
| 2. | Buffer | 10 X | 1X | 2 |
| 3. | MgCl2 | 50 mM | 2mM | 0.8 |
| 4. | dNTPs | 10 mM | 250 µM each | 0.5 |
| 5. | Forward Primer | 10 µM | 0.1 µM | 0.2 |
| 6. | Forward Primer | 10 µM | 0.1 µM | 0.2 |
| 7. | Taq DNA polymerase | - | Optimize in the lab | 0.15 |
| 8. | MQ water | - | - | 14.15 |
| Total | 20 µl | |||
Table1: List of PCR reagents, their working concentration and amount used in PCR.
| S.No. | Steps in PCR Program | Temperature | Time | No. of Cycles |
|---|---|---|---|---|
| 1. | Initial Denaturation | 95 ◦C | 5 min | 1 |
| 2. | Denaturation | 95 ◦C | 45 Sec | 45 |
| Annealing | Optimized to each primer pair | 45 Sec | ||
| Extension | 72 ◦C | 45 Sec | ||
| 3. | Final Extension | 72 ◦C | 4 min | 1 |
| 4. | Hold | 4 ◦C | Infinite | 1 |
Table2: List of different steps and their time duration used in PCR.
Agarose gel electrophoresis
Agarose gel electrophoresis requires the following items: An electrophoresis chamber and power supply. Gel casting trays, which are available in a variety of sizes and composed of UVtransparent plastic. Sample combs, around which molten agarose is poured to form sample wells in the gel. Electrophoresis buffer, usually Tris-borate-EDTA (TBE).
DNA Loading dye or Buffer, which contains glycerol (30%), Xylene cyanol (0.25%) and Bromophenol Blue (0.25%). Glycerol prevents the diffusion of sample while loading of samples into wells whereas tracking dyes help the visual monitoring of electrophoresis. Ethidium bromide, a fluorescent dye used for staining nucleic acids.
Trans-illuminator (an ultraviolet light box), which is used to visualize ethidium bromide-stained DNA in gels. DNA Ladders, DNA fragments of known sizes that are typically generated by restriction enzyme digestion of a plasmid or bacteriophage DNA of known sequence. DNA ladders are used to check the specificity of primers as well as quantitation of amplified products. The different concentrations of agarose used for DNA fragments of various range of length are mentioned in the following Table 3.
| Agarose Concn (% [w/v]) | Range of Linear DNA Molecules (kb) |
|---|---|
| 0.3 | 5-60 |
| 0.6 | 1-20 |
| 0.7 | 0.8-10 |
| 0.9 | 0.5-7 |
| 1.2 | 0.4-6 |
| 1.5 | 0-2-3 |
| 2.0 | 0.1-2 |
Table 3: Agarose concentration and respective DNA fragment size.
Results
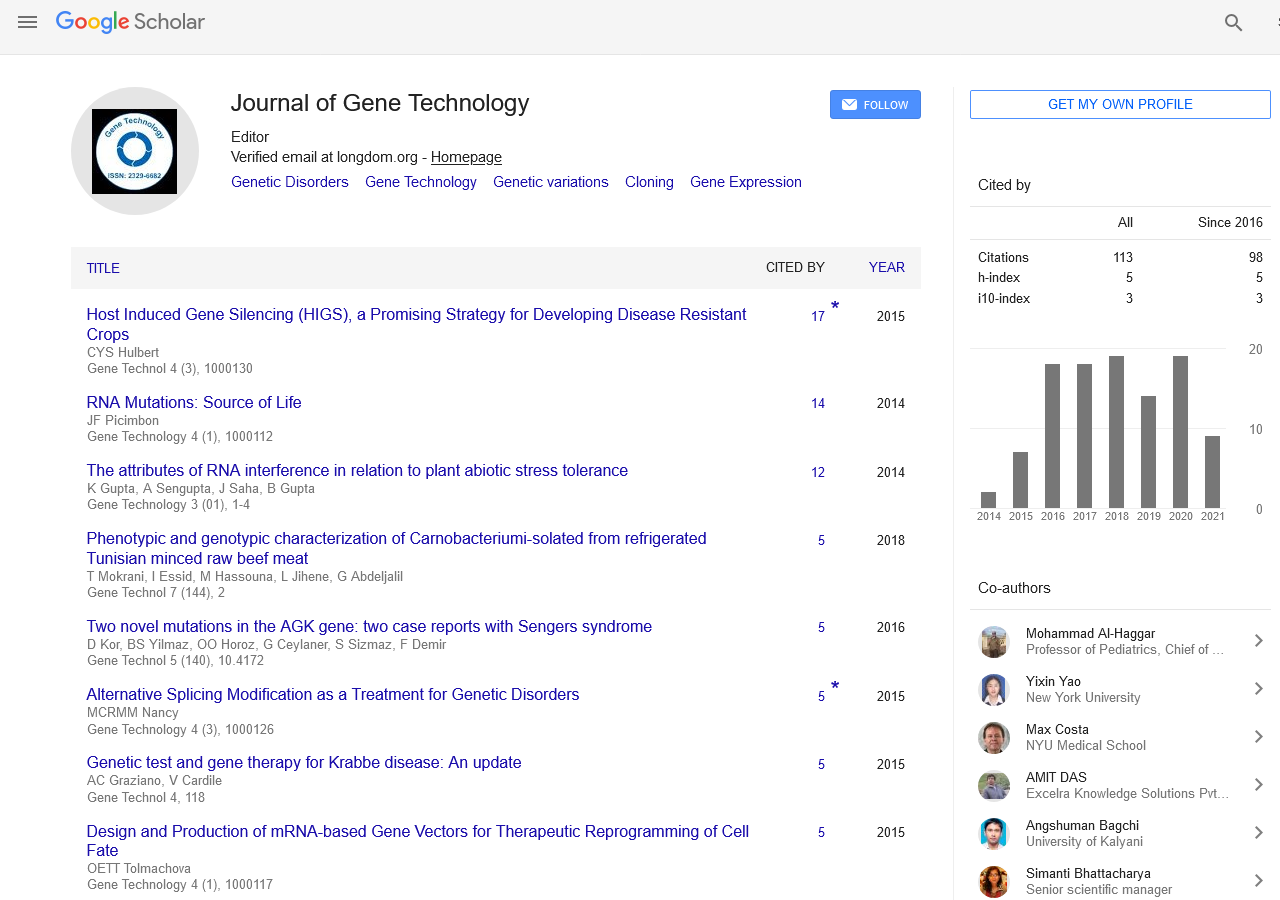
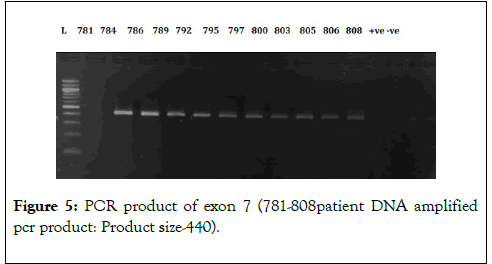
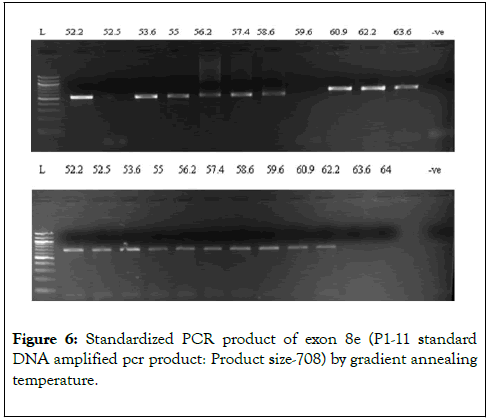
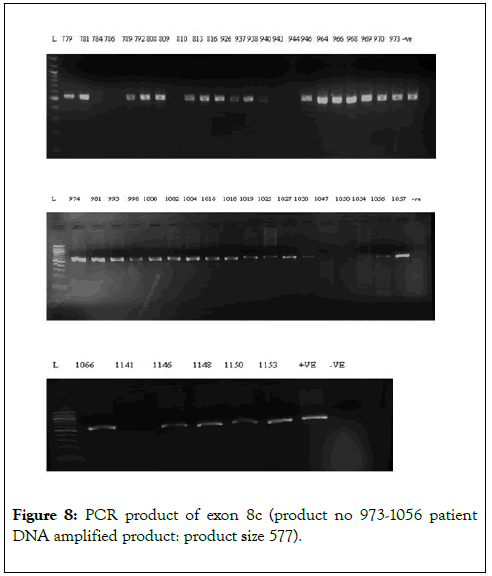
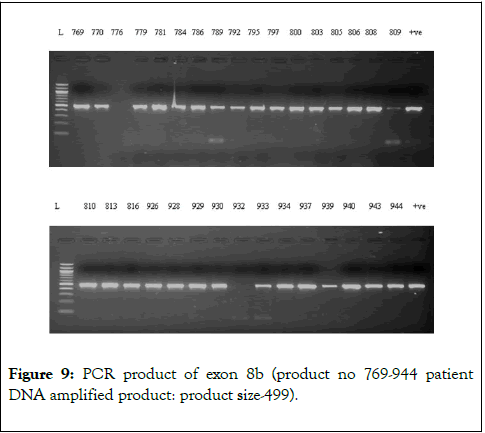
We have chosen FOG2 for the identification of rare variations in CHD patients from Indian population. To evaluate the study we have done the PCR amplification for Exon 7 and Exon 8, since most of the zinc finger domains of FOG2 are localized in these two exons. Since, exon 8 is quite bigger in length is approximately 2491bp. It was divided into 5 different regions. i.e. 8a-8e. For exon 7 we did amplification for 12 samples observed the gel picture result shows in (Figure 5). To find out the exact annealing temperature for 8e, standard DNA used two times to get a constant annealing temperature by gradient and confirmed the temperature for proceeding further work (Figure 6). For exon 8a and 8c we did amplification for 96 samples respectively and observed the gel picture result shows in (Figures 7 and 8). We observed PCR product result for exon 8b used samples (Figure 9).
Figure 5: PCR product of exon 7 (781-808patient DNA amplified pcr product: Product size-440).
Figure 6: Standardized PCR product of exon 8e (P1-11 standard DNA amplified pcr product: Product size-708) by gradient annealing temperature.
Figure 7: Showing the amplified PCR product of exon 8a from CHD patient DNA (product size-559)
Figure 8: PCR product of exon 8c (product no 973-1056 patient DNA amplified product: product size 577).
Figure 9: PCR product of exon 8b (product no 769-944 patient DNA amplified product: product size-499).
Discussion
Congenital heart disease is one of the most common birth defects with leading cause of morbidity and mortality throughout the world. Among several possible causative factors, single nucleotide changes in the key transcription factors are of great importance in contributing the occurrence of CHD.FOG2, a transcriptional cofactor of GATA4, is well known gene which is expressed at early cardiac development in cardiac tissues (Crispino John et al.) [12]. Mutations in the coding region of this gene have been reported as potential causative agent for various CHD phenotypes.
Conclusion
In this study, we have amplified the c-terminal end of protein (exon 7 and exon8) by polymerase chain reaction. Exon 8 was subdivided into 5 different region i.e. Exon8A-8E because it has long coding region. These exons were amplified in different conditions. For example, Exon 8A-B and E were amplified with Platinum taq DNA polymerase without requirement of any PCR enhancers such as DMSO (Dimethylsulfooxide) or betaine. However, Exon 8C was amplified in the presence of DMSO. The annealing temperature for every exons is different. Exon 7 is amplified Taq DNA polymerase. The specificity and quantity of amplified products are checked by the 100bp DNA ladder.
REFERENCES
- Hoffman JI, Kaplan S. The incidence of congenital heart disease. J Am Coll. Cardiol. 2002;39(12):1890-1900.
- Pierpont ME, Basson CT, Benson DW, Gelb BD, Giglia TM, Goldmuntz E, et al. Genetic basis for congenital heart defects:current knowledge: a scientific statement from the American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young. Circulation. 2007;115(23):3015-38.
- Cooper WO, Hernandez-Diaz S, Arbogast PG, Dudley JA, Dyer S, Gideon PS, et al. Major congenital malformations after firsttrimester exposure to ACE inhibitors. N Engl J Med. 2006; 354:2443-2451.
- Bruneau BG. The developmental genetics of congenital heart disease. Nature. 2008;451:943-948.
- Rochais F, Mesbah K, Kelly RG. Signaling Pathways Controlling Second Heart Field Development. Circ Res. 2009;104(8):933-942.
- Schott JJ, Benson DW, Basson CT, et al. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science. 1998;281:108-111.
- Laity JH, Lee BM, Wright PE. Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol. 2001;11(1):39-46.
- Peterkin T, Gibson A, Loose M, Patient R. The roles of GATA-4,-5 and -6 in vertebrate heart development. Semin Cell Dev Biol. 2005;16(1):83-94.
- Pu WT, Ishiwata T, Juraszek AL, Ma Q, Izumo S. GATA4 is a dosage-sensitive regulator of cardiac morphogenesis. Dev Biol. 2004; 275(1): 235-44.
- Rojas A, Kong SW, Agarwal P, Gilliss B, Pu WT, Black BL. GATA4 is a direct transcriptional activator of cyclin D2 and Cdk4 and is required for cardiomyocyte proliferation in anterior heart field-derived myocardium. Mol Cell Biol. 200;28(17):5420-31.
- Manuylov NL, Sergei G. Cardiac expression of Tnnt1 requires the GATA4- FOG2 transcription Complex. The Scientific World. 2009;9:575-587.
- Crispino JD, Lodish MB, Thurberg BL, Litovsky SH, Collins T, Molkentin JD, et al. Proper coronary vascular development and heart morphogenesis depend on interaction of GATA-4 with FOG cofactors. Genes Dev. 2001;15(7):839-844.
Citation: Elangovan B (2020) Mutational analysis of FOG2 gene in patients with congenital Heart disease. Gene Technol. 9:152. DOI: 10.35248/2329-6682.20.9.152
Copyright: © 2020 Elangovan B. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.