PMC/PubMed Indexed Articles
Indexed In
- Academic Journals Database
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- ResearchBible
- Ulrich's Periodicals Directory
- Electronic Journals Library
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- MIAR
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2019) Volume 10, Issue 1
Genetic Mutations in the Papuan Human Mitochondrial Genome: Studies in Gene Control Regions and Gene Coding Using REPLI-g
Yohanis Ngili* and Johnson SiallaganReceived: 12-Nov-2019 Published: 03-Dec-2019
Abstract
Comparative studies of analysis and DNA mutations in Papuan humans and its comparison with several world ethnicities, both in the coding region and the gene control region have been conducted. The objective of this research was to analyze mutation variants in all regions of the complete genome of the human mitochondria by using the G-repli technique for amplification of the mtG, the results of nucleotide sequencing were then compared on several individuals representing some ethnicities in the world. DNA samples were isolated from human tissue and then sequenced using efficient primary pairs to amplify human mtG.
Here, we report that the results of mutation analysis through the mtDNA cloning process can be concluded that variations in the length of the poly-C series in the same sample indicate the presence of mtDNA subpopulations in certain individuals, also known as heteroplasms. This phenomenon is strongly suspected to be the cause of the unreadable sequence of the D-loop HVSI region that has poly-C through the direct sequencing method. This is thought to have occurred because of several subpopulations in different Papuan humans in one sample, which caused the sequencing detector to receive two different fluorescence signals at the same position. This signal difference occurs because of the shifting of the mtDNA nucleotide base due to differences in the length of the poly-C series. This allegation has opened the opportunity for further research on the relationship between unreadable sequences of mtDNA-containing HVSI regions containing poly-C through direct sequencing with variations in the composition of different subpopulations. This concept is important in studying mtDNA mutations associated with diseases (diseaserelated mutations).
Keywords
mtG; Variant; Nucleotide mutation; Papua; Ethnic groups
Introduction
Mitochondria have two kinds of membranes, the outer membrane and the inner membrane. The outer membrane contains a number of transport proteins, called porins, which are able to filter out ions or small molecules of 5 kDa or less. In addition, the outer membrane also contains enzymes involved in lipid biosynthesis and enzymes which play a role in the transport of lipids to the matrix to undergo β-oxidation to produce acetyl CoA. Mitochondria are eukaryotic cell organelles whose role is to produce energy in the form of compounds adenosine triphosphate (ATP) through a highly efficient oxidative phosphorylation reaction [1-3].
Unlike other organelles cell, mitochondria have their own genetic material whose characteristics are different from genetic material in the cell nucleus. The uniqueness of mitochondrial DNA includes a higher mutation rate, which is 10-15 times that of core DNA [4]. In addition mitochondrial DNA is present in large quantities (more than 1000 copies) in each cell, while the core DNA is only two copies [5]. Nuclear DNA is the result of recombination of the DNA of both parents, whereas mitochondrial DNA is only inherited from the mother (maternal inherited) [6]. Mutations that occur in the mitochondrial genome (mitochondrial genome/mtG) can also increase the production of intracellular free radicals, so that the production of free radicals decreases mitochondrial function and will increase the accumulation of oxidation processes in various tissues [7,8].
The sequence and organization system of the human mitochondrial genome was first published by Anderson and his colleagues [9]. Since then, studies studying mtDNA sequences, genetic mutations, and massive mutation comparisons have been carried out. The structure of mtDNA is circular in shape and consists of 16.5 kb. There have been many results of human mitochondrial DNA research with its unique characteristics to be utilized in various scientific disciplines, including studies on evolution, population genetics, bioinformatics, genetic diseases, and forensic medicine [1,10-13].
Material and Methods
Samples are individual blood and oral epithelial cells. Samples were stored in 1.5 mL eppendorf tubes frozen (-20 C). The procedure of isolating mitochondrial DNA from human tissue follows the DNA Purification from Tissues protocol. Tissue samples were weighed using an analytical balance weighing 25 mg (1 mg tissue is equivalent to 0.2-1.2 μg DNA) [14]. The in vitro REPLI-g amplification procedure is by taking 5 μL of the isolated DNA template using the QIAamp DNA Mini Kit [15]. Amplification of mtDNA by PCR in this study uses the 2400 GeneAmpPCR system (Perkin Elmer). The primers for amplification are designed using the Perlprimer program to facilitate simulations for determining annealing temperatures [15].
The amplification reaction of some fragments was catalyzed by the Green DreamTaq PCR Mastermix enzyme containing DreamTaq DNA Polymerase. Mastermix is added with two tracking dyes and ballast reagents that function as direct loading for electrophoresis, so when electrophoresis does not need to be added to loading dye. The PCR process was carried out with an Automatic thermal cycler machine of 30 cycles. The initial stage of the PCR process is the initial denaturation stage carried out at a temperature of 95°C for 3 minutes, then entered into the PCR cycle program with each cycle consisting of 3 stages: the denaturation stage which is carried out at a temperature of 95°C for 30 sec, the annealing stage performed at a temperature of 50°C for 30 sec and the extension or polymerization at a temperature of 72°C for 60 sec. At the end of all cycles an additional polymerization process was carried out at 72°C for 10 min [16,17].
PCR products were detected by electrophoresis on 1.2% agarose gel (w/v) using a mini subTM DNA electrophoresis. The electrophoresis process was carried out in a 1x TAE buffer as a medium conducting current at 75 volts for 45 minutes. Electrophoresis results were visualized with a series of 9814-312 nm UV lamps (Cole Parmer). For determining the mtG nucleotide sequence PCR primers and several internal primers are used. The multiple alignments of human complete mtG nucleotide sequences were analyzed with the help of DNAstar software program. MTG analysis was performed using the EditSeq, Seqman, and MegAlign programs [18-20].
Results and Discussion
Preparation of DNA templates for PCR begins with sampling of blood cells for samples of ethnic Papuan human individuals. Furthermore cell lysis is performed to obtain mitochondrial DNA using a standard procedure without prior purification [20]. The extracted DNA is a template for the Polymerase Chain Reaction (PCR) process (Figure 1) shows the 1 kb fragment of the D-loop region obtained by amplification of human mtDNA in vitro using a pair of Mfor and Mrev primers. DNA detection results show that the sample gives an amplification of about 1 kb in size as indicated by the existence of a band located between the 1000 band and the 1500 pb band. This result is as expected, which amplifies the D-loop region consisting of HVS1 and HVS2 with a length of about 1 kb.
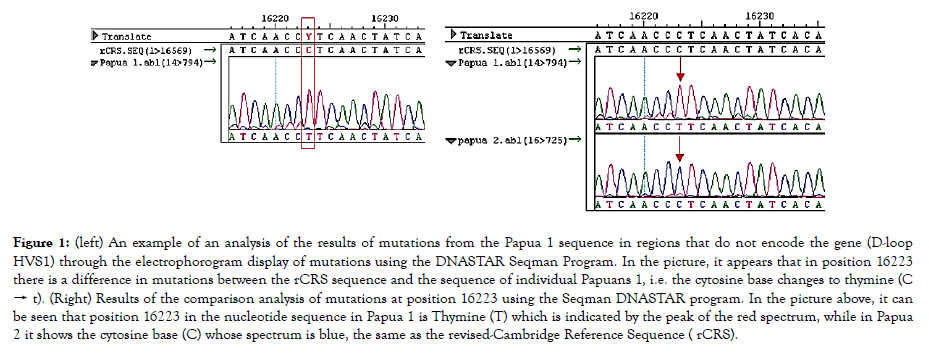
Figure 1. (left) An example of an analysis of the results of mutations from the Papua 1 sequence in regions that do not encode the gene (D-loop HVS1) through the electrophorogram display of mutations using the DNASTAR Seqman Program. In the picture, it appears that in position 16223 there is a difference in mutations between the rCRS sequence and the sequence of individual Papuans 1, i.e. the cytosine base changes to thymine (C → t). (Right) Results of the comparison analysis of mutations at position 16223 using the Seqman DNASTAR program. In the picture above, it can be seen that position 16223 in the nucleotide sequence in Papua 1 is Thymine (T) which is indicated by the peak of the red spectrum, while in Papua 2 it shows the cytosine base (C) whose spectrum is blue, the same as the revised-Cambridge Reference Sequence ( rCRS).
The results of DNA amplification were carried out using the repliG technique to amplify ten mtG fragments. The results of this amplification produce bright, consistent DNA bands for each PCR result. The PCR process using REPLI-g can minimize DNA contamination during the isolation process and PCR. Electrophoresis results from human DNA samples that have been successfully analyzed by PCR, and then electrophoresed for kidney tissue.
Data of human nucleotide sequences from the tissues were analyzed using DNASTAR program in the form of editseq and ABI files. The sequence data is then carried out homology analysis with the nucleotide sequence rCRS using DNAstar program namely Megalign and Seqman, which is to determine the presence of mtG nucleotide mutations. The results of the mtG mutation study as a whole are concluded that the mutations that occur in humans are generally transitional nucleotide mutations. Causes of transitional nucleotide mutations include nitric acid, base pairing errors, and mutagenic base analogues [16,17]. Based on the results of the analysis, substitution mutations could be returned to normal conditions by conducting a transition or reverse transversion process. Hence the substitution effect is not permanent (Figure 2).

Figure 2. The results of the alignment of human mtDNA with rCRS in the gene coding region. One example of secondary data analysis is mtG at a certain position, where there are successive changes in G→a and T → c in Papuan Individuals. Individual C sequences from GenBank were analyzed in silico using the Megalign program on rCRS.
Nucleotide data from the analysis revealed several mutations in rCRS. Observation of mutations shows that the rate of mitochondrial DNA mutations turns out to be different between one individual and another individual. Accumulation of mtDNA that carries a mutation will affect the tissue's oxidative energy metabolic capacity, so this capacity will decrease with age. The effect of this reduction is proposed to be manifested especially in tissues that require a lot of ATP for its function, and is seen as a decreased ability of the heart muscle and brain function. Much of the mtG mutation data related to disease and comparison between ethnicities has been entered into the MITOMAP database.
The observations showed that the variants of human mitochondrial DNA mutations turned out to be different between one individual and another as happened in the number and position of mutations in both A individuals and B individuals as well as C-N individual comparisons. This is because the mtDNA mutation is inherited only in line with maternal lineage (maternal lineage), while the individuals analyzed are from different breeds and various ethnicities. After the database of variations of human mtG nucleotides against rCRS has been completed, the next step is to conduct an analysis of the size of human mtG nucleotides. The size of mtG analyzed ranged between 16558-16574 bp from 301 GenBank samples. The most mtG size is in accordance with the length of base pairs first proposed by Anderson and his colleagues, namely 16569 bp, as many as 113 samples. The next highest number of base pairs in a row, size 16570 bp (97 samples), 16571 bp (44 samples), 16572 bp (10 samples), 16574 bp (3 samples), 16573 bp (2 samples), 16558 bp and 16559 bp (1 sample each). This is done to observe the mutation rate in the mtG nucleotide sequence of each individual human; because it has been known beforehand that the mutation rate that occurs in mtG varies per individual who is not in line with maternal descent.
The number of nucleotide variants (mutations against rCRS) in each sample turned out to range from 12 (the smallest number of mutations) to 42 (the largest number of mutations) from a total of 301 human mtG samples originating from GenBank. The highest number of mutations is 15 mutations found in 42 individuals or 13.95%. The next five most mutations were 17 mutations (39 individuals or 12.95%), 16 mutations (37 individuals or 12.29%), 18 mutations (36 individuals or 11.96%), 14 mutations (20 individuals or 6.64%), and 19 mutation variants (18 individuals or 5.96%). The MITOMAP database provides a variety of fairly informative data, such as mutations that have been reported, diseases related to mutations in mitochondrial DNA, researchers who report, data related to the origin of samples (ethnicity), and other information. Information about mtDNA mutations associated with diseases is very useful for the medical world because it makes it easier to search for information about mutations related to mitochondrial diseases. This is of course very important for the future revolution of the medical world because it is related to beautique medicine.
Conclusions
The results of mutation analysis showed that there were point mutations in several mtG region fragments with different proportions of mutations. Most mutations outside the D-loop regions are in the ATP6 region. Analysis of the variability of the nucleotide sequences of Papuan human individual samples provides information on the identity of mutations that vary greatly in position, number and type of mutation that occurs. The emergence of variability in position, number and type of mutations in Papuan individuals indicates that the mutations that occur are not specific to certain populations such as the Federal Bareau of Investigation (FBI) mtDNA Population Database. While the results of the analysis in the gene coding region provide enough variability of mutations between tribes in Papua which is quite high. This study is quite interesting to examine more deeply in bioetnoanthropological studies.
REFERENCES
- Choo-Kang ATW, Lynn S, Taylor GA, Daly ME, Sihota SS, Wardell TM, et al. Defining the importance of mitochondrial gene defects in Maternally Inherited Diabetes by Sequencing the Entire Mitochondrial Genome. Diabetes. 2002;51(7):2317-2320.
- Albert B, Bray DJ, Watson JD, Raff JM, Roberts K. Molecular biology of the cell. Garland Publishing Inc. New York. USA. 1994. pp: 146.
- Ballana E, Morales E, Rabionet R, Montserrat B, Ventayol M, Bravo O, et al. Mitochondrial 12S rRNA gene mutations affect RNA secondary structure and lead to variable penetrance in hearing impairment. Biochem Biophys Res Commun. 2006;341(4):950-957.
- Rebecca LC, Stoneking M, Wilson AC. Mitochondrial DNA and human evolution. Nature. 1987;31-36.
- Elliot HR, Samuels DC, Eden JA, Relton CL, Chinnery PF. Pathogenic mitochondrial DNA mutations are common in the general population. Am J Hum Genet. 2008;83(2):254-260.
- Flaherty KR, Wald J, Weisman IM, Zeballos RJ, Schork MA, Blaivas M, et al. Unexplained exertional limitation: Characterization of patients with a mitochondrial myopathy. Am J Respir Crit Care Med. 2001;164(3):425-432.
- Ingman M, Gyllensten U. Analysis of the Complete Human mtDNA Genome: Methodology and Inferences for Human Evolution. Jour of Heredi. 2001;92(6):454-461.
- Kogelnik AM, Lott MT, Brown MD, Navathe SB, Wallace DC. MITOMAP: A human mitochondrial genome database-1998 Update. Nucleic Acids Res. 1998;26(1):112-115.
- Lertrit P, Noer AS, Jean-Francois MJ, Kapsa R, Dennett X, Thyagarajan D, et al. A new disease-related mutation for mitochondrial encephalopathy lactic acidosis and strokelike episodes (MELAS) syndrome affects the ND4 subunit of the respiratory complex I. Am J Hum Genet. 1992;51(3):457-468.
- MarzukiA S, Noer AS, Lertrit P, Thyagarajan D, Kapsa R, Utthanaphol E, et al. Normal variants of human mitochondrial DNA and translation products: The building of a reference data base. 1991;88(2):139-145.
- Meissner C, von Wurmb N, Schimansky B, Oechmichen M. Estimation of age at death based on quantitation of the 4977-bp deletion of human mitochondrial DNA in skeletal muscle. Foren Sci Int. 1999;115-124.
- MITOMAP, 2011, www.mitomap.org,
- Moraes CT, Kenyon L, Hao H. Mechanisms of human mitochondrial DNA maintenance: The determining role of primary sequence and length over function. Mol Biol Cell. 1999;10(10):3345-3356.
- Newton CR, Graham A. PCR-Introduction to Biotechniques. BIOS Scientific Publishers. Mortimer Street London, United Kingdom. 1994.
- Ngili Y, Syukriani YF, Ahmad AS, Noer AS, Natalia D, Syah MD. Variants analysis of human mitochondrial genome mutation: Study on indonesian human tissues. International Journal of Chem Tech Research. 2012;4(2):720-728.
- Noer AS, Sudoyo H, Lertrit P, Thyagarajan D, Utthanaphol P, Kapsa R, et al. A tRNA(Lys) mutation in the mtDNA is the causal genetic lesion underlying myoclonic epilepsy and ragged-red fiber (MERRF) syndrome. Am J Hum Genet. 1991;49(4):715-722.
- Nomiyama T, Tanaka Y, Hattori N, Nishimaki K, Nagasaka K, Kawamori R, et al. Accumulation of somatic mutation in mitochondrial DNA extracted from peripheral blood cells in diabetic patients. Diabetologia. 2002;45(11):1577-1583.
- Helen AL, Tuppen, Emma LB, Turnbull DM, Taylo RW. Mitochondrial DNA mutations and human disease. Biochimica et Biophysica Acta (BBA)-Bioenergetics. 2010;1792(2):113-128.
- Wallace DC, Singh G, Lott MT, Hodge JA, Schurr TG, Lezza AM, et al. Mitochondrial DNA mutation associated with Leber's hereditary optic neuropathy. Science. 1988;242(4884):1427-1430.
- Wei YH, Lee HC. Mitochondrial DNA mutations and oxidative stress in mitochondrial diseases. Adv Clin Chem. 2003;37:83-128.
Citation: Ngili Y, Siallagan J (2019) Genetic Mutations in the Papuan Human Mitochondrial Genome: Studies in Gene Control Regions and Gene Coding Using REPLI-g. J Data Mining Genomics Proteomics. 10:218. DOI: 10.35248/ 2153-0602.19.10.218
Copyright: © 2019 Ngili Y, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.


