PMC/PubMed Indexed Articles
Indexed In
- Academic Journals Database
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- China National Knowledge Infrastructure (CNKI)
- Scimago
- Ulrich's Periodicals Directory
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- MIAR
- University Grants Commission
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2021) Volume 0, Issue 0
Galectin Antagonist use in Mild Cases of SARS-CoV-2: Pilot Feasibility Randomised, Open Label, Controlled Trial
Alben Sigamani1*, Mathu Ruthra1, Sudhishma1, Samarth Shetty2, Madhavi2, Anup Chugani3, Hana Chen-Walden4, David Platt4 and Thomas Kutty52Mazumdar Shaw Medical Center, Narayana Health, Bangalore, India
3Department of Molecular Biology, Medgenome Labs, Bangalore, Karnataka, India
4Pharmalectin Inc, Boston, USA
5Baylor Scott & White Health, Texas, USA
Received: 08-Dec-2020 Published: 30-Dec-2020, DOI: 10.35248/2157-7560.21.S10.003
Abstract
Importance: Novel SARS-CoV-2 virus has infected nearly 100 million people across the world and is highly contagious. There is a need for a novel mechanism to block viral entry and stop its replication.
Background: Spike protein N Terminal Domain (NTD) of the novel SARS-CoV-2 is essential for viral entry and replication in human cell. Thus the S1 NTD of human coronavirus family, which is similar to a galectin binding site-human galactose binding lectins, is a potential novel target for early treatment in COVID-19.
Objectives: To study the feasibility of performing a definitive trial of using galectin antagonist–Prolectin-M as treatment for mild, symptomatic, rRT-PCR positive, COVID-19.
Main outcomes and measures: Cycle threshold (Ct) value is number of cycles needed to express fluorescence, on real time reverse transcriptase polymerase chain reaction. Ct values expressed for RNA polymerase (Rd/RP) gene+Nucleocapsid gene and the small envelope (E) genes determine infectivity of the individual. A digital droplet PCR based estimation of the Nucleocapid genes (N1+N2) in absolute copies/μL determines active viral replication.
Design and intervention: Pilot Feasibility Randomised Controlled Open-Label, parallel arm, study. Oral tablets of Prolectin-M were administered along with the best practice, Standard of Care (SoC) and compared against SoC. Voluntarily, consenting individuals, age>18 years, and able to provide frequent nasopharyngeal and oropharyngeal swabs were randomly allocated by REDCap software.
The intervention, Prolectin-M was administered as a multi dose regime of 4 gram tablets. Each tablet contained 2 grams of (1-6)-Alpha-D-mannopyranosil (galactomannan) mixed with 2 grams of dietary fibre. Each participant took a single chewable tablet every hour, to a maximum of 10 hours in a day. Tablets were administered only during the daytime, for total of 5 days.
Results: This pilot trial demonstrated the feasibility to recruit and randomize participants. By day 7, following treatment with Prolectin-M, Ct value of Rd/Rp+N gene increased by16.41 points, 95% (CI 0.3527 to 32.48, p=0.047). Similarly, small envelope (E) gene also increased by 17.75 points (95% CI,-0.1321 to 35.63, p=0.05). The expression of N1, N2 genes went below detectable thresholds by day 3 (Mann Whitney U=0.000, p<0.029).
rRT-PCR testing done in the clinic on day 1, 7, and 14 had 3 participants (60%) turn negative by day 7 and all turned negative by day 14 and stayed negative until day 28. In the SoC group 2 participants had zero detectable viral loads at baseline, 2 participants tested negative on day 14, and the last participant remained positive on day 28. There were no serious adverse events, and all participants were clinically asymptomatic before day 28 with reactive immunoglobulin G (IgG).
Trial relevance: This pilot study proves that it is feasible and safe to perform a trial using a Galectin antagonist in COVID-19. This is a novel mechanism for blocking viral entry and its subsequent replication.
Keywords
Vaccine; RNA; COVID-19; Galectin; Immunoglobulin
Introduction
SARS-CoV-2 has infected over 80 million people worldwide and is responsible for over 1.1 million deaths [1]. Several treatments have been authorized by regulators around the globe and none of them have been able to lower its infectivity [2]. SARS-CoV-2, a new member of the coronaviruses known to infect humans [3], is a single-stranded RNA-enveloped virus, with a large number of glycosylated S proteins covering its surface [4]. These proteins mediate viral cell entry following a S protein binding on host cell surface [5].
During viral infection, the target cells activate the S protein, by cleaving it into S1 and S2 subunits [6]. The S1 subunit is further divided into a N-terminal domain (NTD) and a C-terminal domain (CTD). The CTD is known to bind the human angiotensin converting enzyme-2 (ACE-2) receptors while NTD seeks gangliosides on cell surface to stabilize cell adhesion [7,8].
SARS-CoV-2 belong to betacoronaviridae family, which includes mouse hepatitis coronavirus (MHV). The S1-NTD structure, described in the mouse hepatitis coronavirus (MHV), demonstrates a similar structural fold as the human galactose binding lectins (galectins) [9]. Galectins are carbohydrate-binding proteins, involved in many physiological functions, such as inflammation, immune responses, cell migration, autophagy and signaling [10]. They are also involved immunogenicity, i.e. cell recognition, between human cells and infective pathogens; viruses, bacteria, and parasites [11].
Prolectin – M is an orally administered polysaccharide (galactomannan). Polysaccharides competitively bind to the N-terminal tail of human galectin-3 through a proline isomerization [12]. Galetin-3 (Gal-3) is 1 among the 15 galectins described in humans and also a ubiquitous human galectin expressed in various disease pathogenesis pathways [13,14]. The objective of this trial is to demonstrate the feasibility of a large randomised controlled clinical trial, and a possible mechanism of action for galectin antagonists as treatment in COVID-19.
Viral culture studies of SARS-CoV-2 [15] indicate, that date of onset of symptoms and cycle threshold levels relate to infectivity; a cycle threshold of <25 is considered to be infectious [16,17]. A Cycle threshold (Ct) value is the number of replication cycles required for a signal of reverse transcription polymerase chain reaction (rRTPCR) product, to cross a determined threshold.
Most rRT-PCR approved kits, report Ct values for the RNA dependent RNA polymerase (Rd/Rp), envelope (E) protein and nucleocapsid genes. All three genes are reported together for higher sensitivity. The droplet digital PCR measures expression of only nucleocapsid genes (N1 and N2) as an absolute number in copies/uL [18]. We hypothesized that blocking S1 NTD using a galectin antagonist can affect viral replication in nasopharyngeal, oropharyngeal, gastrointestinal tract, and potentially clear the virus from the person to make them non-infectious in a shorter time [19].
For this pilot study we considered a rising number in the Ct values, along with a reduction in the number of copies/µL of the nucleocapsid gene as a measure of our endpoint. However, in future clinical trials Prolectin-M should be tested using an endpoint that measures the effect on time to recovery in days as well as the reduction of infectivity [20,21]. A galectin antagonist could treat COVID-19 patients early in the disease pathogenesis and prevent spread of SARS-CoV-2 within the community [22].
Methods
Trial design and oversight
This investigation is a pilot and feasibility open-label randomized controlled trial. The trial protocol appears in supplement 1. A total of 10 subjects were identified between September 15th and 19th, 2020. Study data was collected and managed using REDCap electronic data capture tools hosted at [Narayana Hrudayalaya Limited Bangalore] [23,24].
REDCap (Research Electronic Data Capture) is a secure, web-based software platform designed to support data capture for research studies, providing 1) an intuitive interface for validated data capture; 2) audit trails for tracking data manipulation and export procedures; 3) automated export procedures for seamless data downloads to common statistical packages; and 4) procedures for data integration and interoperability with external sources.
This study followed the Consolidated Standards of Reporting Trials (CONSORT) 2010 extension to randomized pilot and feasibility trials [25]. The study was registered at clinicaltrials. gov (NCT04512027) and the Clinical Trials Registry of India (CTRI/2020/09/027833). Study protocol and all documents were reviewed by Institutional Ethics Committee – Narayana Health and approved (NHMEC Ref. No. S44/2020), on 31st of August 2020. The study was conducted in compliance with the Declaration of Helsinki, the Good Clinical Practice guidelines, and local regulatory requirements.
Before enrolment and allocation to the study arms, informed consent for participation in the study was obtained in writing from all participants. The participant also signed a form, declaring that they received the correct information and gave informed consent for voluntary participation in the trial, before being randomized.
Participants
This single center trial enrolled hospitalized, eligible, and consenting participants between the ages of 18 and 45. All patients had to have an instrumental diagnosis for COVID-19; a positive rRT-PCR for SARS-CoV-2 obtained from an outpatient collection of nasopharyngeal swabs. Other inclusion criteria were the presence of symptoms that were not older than 72 hours and an ability to provide consent to undergo repeated collection of throat and nasal swabs over the 7-day period. Samples were collected pre randomization, during drug administration on days 3 and 5 and 7.
Exclusion criteria included oxygen saturation at admission ≤ 96%, high temperature ≥ 1000F (≥ 37.5°C) not controlled on oral doses of acetaminophen, known history of diabetes on oral medications or insulin therapy or interleukin-6 levels ≥ 3 times of laboratory reference range and/or significantly elevated levels of CRP, serum ferritin or d-dimer or a Lymphocyte/monocyte ratio ≤ 3 or neutrophil/lymphocyte ratio ≥ 5 or platelet count ≤ 150,000 cells per microliter. Previously tested positive and recovered COVID-19 were excluded. Also participants on any chronic medications for more than 4 weeks before randomization or active malignancy or having any co-morbidity that increases risk of rapid disease progression were excluded.
Objective
The primary aim was to evaluate the efficacy, safety and feasibility of administering Prolectin-M versus the SoC for 5 days. The primary endpoint was a change in absolute count of Nucleocapsid gene and a rising Ct value, estimated from serial samples of RNA extracted from a nasopharyngeal swab. The swab was collected in all participants on days 1 (day of randomisation), 3, 5, and 7. Clinical progression was estimated on a 7-point scale recorded on days 7, 21, and 28. A 2-point change was defined as clinical progression.
7-point severity score (ordinal scale):
• Not hospitalized, no limitations on activities.
• Not hospitalized, limitation on activities.
• Hospitalized, not requiring supplemental oxygen.
• Hospitalized, requiring supplemental oxygen.
• Hospitalized, on non-invasive ventilation or high flow oxygen devices.
• Hospitalized, on invasive mechanical ventilation or extracorporeal membrane oxygenation (ECMO).
• Death
Randomization and treatments
Participants were randomly assigned in a 1:1 ratio to receive a 4 gram tablet of Prolectin-M; a (1-6)-Alpha-D-Mannopyranose, and SoC (Treatment group) or SoC (Control group) via a web-based secure centralized system (REDCap). An independent statistician provided a computer-generated assignment randomization list and blocked with varying block sizes unknown to the investigators.
Prolectin-M was administered orally once every hour up to a maximum dose through the day of 40 grams or 10 tablets a day. The intention was to mimic the viral replication cycle of 8 – 10 hours [26] and also to ensure that the participant is consuming the tablets during the day under supervision of a research nurse. Each subject was encouraged to keep the tablet in their mouth for 1-2 minutes before it dissolved and swallowed. During a mealtime, breakfast, lunch, tea, and dinner, the subject had to wait for 30 minutes after the last meal before taking the next tablet. This was to avoid any potential drop in blood glucose as the tablets could block absorption of carbohydrates consumed in the meal.
Procedures
Patient follow up was maintained for 28 days from the day of randomisation. A sample was considered negative when no Ct value was determined, and no amplification curve was observed or if the Ct value was >29 for all three targets.
A nasopharyngeal/oropharyngeal swab was transported to the research laboratory for RNA extraction in a viral transport media (#MG20VTM-3, MagGenome). All RNA extractions were carried out using the QIAamp Viral RNA mini kit (#52904, Qiagen) following the standard instructions as per the kit protocol.
For each blinded sample the Ct value for Rd/Rp and N, E gene, genes were reported.
rRT PCR: The extracted RNA was analysed on a rRT PCR platform: TRUPCR® SARS-CoV-2 RT qPCR KITV2 (#3B3043B Black Bio Biotech) SARS-CoV-2. To determine the efficiency of the PCR, Ct values obtained from a series of 5 template DNA dilutions of at least 3 different samples were graphed on the y-axis versus the log of the dilution on the x-axis. The Ct values assumed by the following equation were employed to calculate the logarithm of the recombinant gene copy numbers from: Ct=slope × log (Gene Copy Number)+1 where I in the formula acts as the intercept of standard curve [27].
Droplet Digital PCR (DDPCR): The RNA was also analysed on the Droplet Digital PCR (BioRad, USA). This platform is FDA approved for emergency use authorization in COVID-19 [18]. The test is a partition-based endpoint single well RT-PCR test. The DDPCR was combined with rRT PCR because of its higher sensitivity and precision in low viral abundance samples and for the ability to provide absolute copy numbers and any resistance to inhibition often seen in rRT-PCR testing.
The analytic sensitivity was calculated as 0.260 cp/µl to 0.351 cp/ µl (cp=copies) for genetic markers, SARS-CoV-2 N1 and N2 genes, and an internal control human RPP30 gene. The laboratory remained blind to treatment allocation throughout the analysis. On day 28, all participants were tested for serum antibodies against SARS-CoV-2 Immunoglobulin G (IgG) titres (values ≥ 1.00 as reactive) using VITROS® COVID-19 antibody tests [28].
Adverse events were recorded from time of signature of informed consent and graded according to the Common Terminology Criteria for Adverse Events, version 5.0 [29]. Causality was assessed by the investigators for any serious adverse events.
Statistical analysis
Only 10 subjects were randomised and no formal sample estimation was done. Ct values and absolute copy numbers were compared using parametric, unpaired repeated t-test with Welch’s correction or non-parametric Mann Whitney U test. A two-tailed, p<0.05, was considered statistically significant.
Results
All 10 participants were included for final analysis. All participants consented and were randomised within 2 days of their symptom onset. All patients allocated to treatment arm received their medications. Demographic and baseline clinical characteristics are summarized in (Table 1). 8/10 participants were female with a median (range) age of 27.5 (39,25) years. In the control arm 3 patients did not have detectable viral RNA, when tested in the research lab.
| Characteristic | Treated | Control |
|---|---|---|
| Number | 5 | 5 |
| Age | 28.4 ± 3.195 | 29.1 ± 4.53 |
| Sex (Female) | 4 | 4 |
| rRT PCR confirmed sars-CoV-2 infection | 5 | 5 |
| Time from symptom onset to randomization (median) | 1.80 days | 2 days |
| WHO –CPS score (0-10)=4 | Score 2 (5) | Score 2 (5) |
| 7-point ordinal scale | 3 | 3 |
| Co– existing conditions | Nil | Nil |
| Laboratory values | ||
| CRP, mg/L | 1.13 ± 0.800.80 | 4.93 ± 6.546.54 |
| D – Dimer, µg/L | 84.8 ± 78.25 | 95.60 ± 83.73 |
| Neutrophil count | 63 ± 15.24 | 53.82 ± 9.19 |
| Lymphocyte count | 28.89 | 33.78 |
| ± 10.69 | ± 6.6 | |
| 0.691 ± 0.557 | 0.617 ± 0.17 | |
| Lymphocyte to monocyte ratio | 0.41 ± 0.91 | 0.38 ± 0.23 |
| RDW | 14.58 | 16.69 |
| ± 1.9 | ± 2.89 | |
| 13.24 | 12.27 | |
| Hemoglobin | ||
| ± 0.2 | ± 0.8 | |
| ALT/ SGPT | 19 ± 8.98 | 16 ± 5.33 |
| AST / SGOT | 29 ± 8.04 | 25.6 ± 3.43 |
| Creatinine | 0.66 ± 0.11 | 0.85 ± 0.26 |
| Ferritin | 27.96 ± 36.69 | 13.30 ± 7.47 |
| LDH | 219 ± 39.96 | 197 ± 3 6.63 |
| Cycle thresholds | N=5 | N=2 |
| Rd/Rp | 21.306 | 19.549 |
| + Nucleocapsid gene | ||
| Envelope gene | 23.126 | 22.0895 |
Table 1: Baseline characteristics.
Table 2 shows changes that occurred over the follow up period. All treated participants showed rising Ct values from day 3. Difference in mean change in Ct values for the Rd/Rp and N genes, between treated and control group, was 16.41 (95% CI-0.3527 to 32.48, p=0.047, Welch’s t test, two tailed). Difference in Ct values of E gene, 17.75 (95% CI, 0.1321 to 35.63, p=0.05, unpaired t-test with Welch’s correction, 2 tailed).
| Characteristics | N | Treated | N | Control | Difference between two groups |
|---|---|---|---|---|---|
| Change from baseline SARS-CoV-2 status, Day 7 | 5 | 3/5 (60%) negative | 5 | No change | Yes |
| Day 28-Serum Ig G (reactive) | 5 | (All 5 reactive) | 5 | 3 (reactive) | Yes |
| 2 (Non-reactive) | |||||
| Reactive (â?¥1.00) Serum IgG antibody titres on day 28 | 5 | 9.08 (8.68, 10.1) | 3 | 4.58 (0.00, 5.68) | Yes |
| Median (IQR) | |||||
| Change in 7-point ordinal scale day 14 | 5 | All scored 1 | 5 | 3 scored 1 | Yes |
| 2 scored 2 restricted as they were positive | |||||
| Change in 7- point ordinal scale day 21 | 5 | No change from last assessment | 5 | Yes â?? 2 to 1 | No |
| Change in 7- point ordinal scale day 28 | 5 | No change from last assessment | 5 | No change from last assessment | No |
| Cycle threshold on day 7 â?? mean increase from baseline | |||||
| Rd/Rp | 5 | 25.86 | 5 | 9.446 | Yes, 16.41 |
| + Nucleocapsid gene | 95% CI, 0.3527 to 32.48, p=0.047 | ||||
| Envelope gene | 5 | 28.27 | 5 | 10.52 | Yes, 17.75 |
| 95% CI, 0.1321 to 35.63, p=0.05 |
Table 2: Follow up, changes seen after randomization.
Figures 1A and 1B show estimation plots for differences seen in Ct values. Figure 2 shows the change in copies/µL of the nucleocapsid (N1+N2) gene. A Mann Whitney test indicated that the drop in copy numbers among the treated participants was significant compared to the control group; U=0.000, p<0.029. Individual patient wise change in absolute copy numbers is shown in supplementary figure panels.
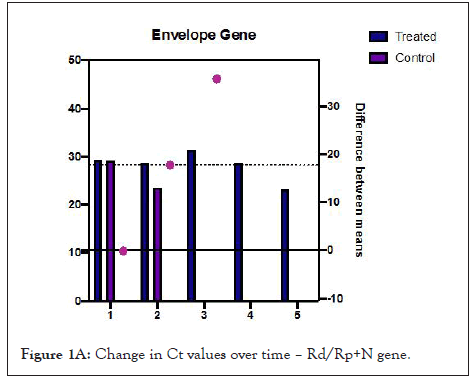
Figure 1A: Change in Ct values over time – Rd/Rp+N gene.
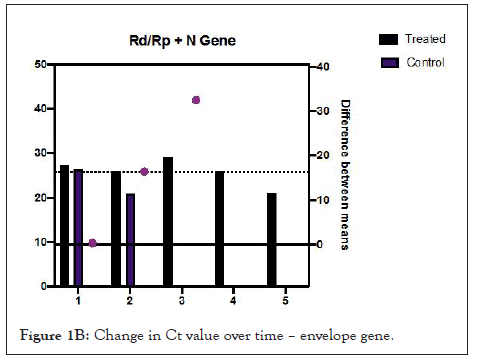
Figure 1B: Change in Ct value over time – envelope gene.
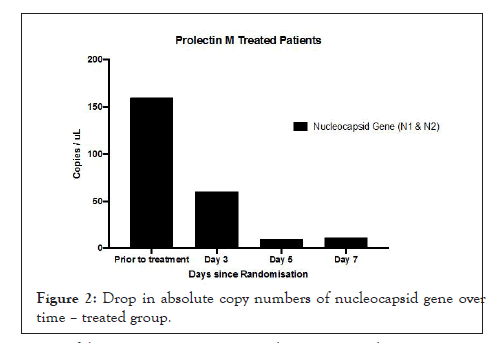
Figure 2: Drop in absolute copy numbers of nucleocapsid gene over time – treated group.
rRT-PCR testing done in the clinic on day 1, 7, and 14 and had 3 participants (60%) turn negative by day 7 and all turned negative by day 14 and stayed negative until day 28. In the SoC group 2 participants had zero detectable viral loads at baseline, 2 participants tested negative on day 14, and the last participant tested remained positive on day 28.
None of the participants experienced any serious adverse event. One participant in the treatment arm had Grade 1 diarrheal episode (CTAE ver. 5.0). No intervention was need, but Prolectin-M was reduced to 5 tablets per day. All participants were fit for discharge on day 14 and with no restrictions except for 2 subjects in the control arm, who tested positive even on day 14. No one had re admission when followed up at day 21 and 28.
Discussion
This is the first ever reported randomised controlled trial of a galectin antagonist against SARS-CoV-2. Blocking N terminal domain (NTD) of S1 subunit, present on spike protein, is seen to have significantly lowered viral gene expression. All five participants in treatment group demonstrated a rapid drop by day 3 in copies/ µL of Nucleocapsid protein gene.
A higher Ct value correlates with lower risk of infectiousness, as reported from a cohort in England{ref}, between January to May 2020. There was rise in Ct>29, for genes, Rd/Rp, N and E genes in the treated group. However, participants in control group (001 and 007), remained potentially infective even on day 7 and day 14.
The detection of anti-spike protein IgG on day 28, is an expression of humoral response against the virus [30]. The N protein is highly immunogenic compared to the S protein and in the absence of glycosylation sites on it results in N-specific neutralizing antibody production early in the stage of acute infection. All participants in the active arm of the trial had a reactive IgG. This trial achieved its primary objective of demonstrating the effect of Prolectin-M on lowering viral infectiousness. The method used to recruit and randomize patients, enabled safe administration of the oral drug which demonstrated an ability to block viral replication (Figure 3).
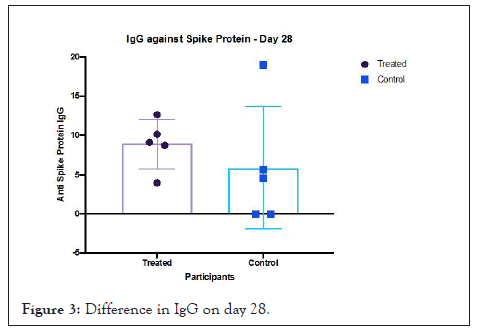
Figure 3: Difference in IgG on day 28.
Strengths of this study are randomisation, concealed allocation, and one hundred percent compliance to treatment, and all procedures in the protocol. There was no incidence of any serious adverse events. A higher cut off value for either cycle threshold or copy numbers on ddPCR as an eligibility criterion could have demonstrated a more significant treatment effect. Hence future trials will need to use either a pre-determined lower (<16) baseline Ct value or a higher copy number for ddPCR.
Galectin antagonists can potentially prevent SARS-CoV-2 entry into human cells [19]. A larger clinical trial will give us the evidence needed to understand its true clinical benefit. Due to the excellent safety profile of galectin antagonists, additional methods of delivery, such as an intravenous, or subcutaneous injection should be examined for a reduction in the number of systemically expressed copies of the virus. In addition, Prolectin-M could be very effective as a prophylactic to stop the community spread of the disease. The results from this pilot and feasibility trial sets the stage for a definitive large randomised controlled clinical trial using galactomannans. as Galectin antagonists have a potential role as a Post Infection Immunisation (PII) Lowering the basic Reproduction number (R0) can reduce SARS-CoV-2 contagiousnes and transmissibility, while protecting the individual from serious COVID-19.
Acknowledgement
There are no acknowledgements to be mentioned.
Source of Funding: Pharmalectin Inc contracted the trial to a Contract Research Organisation – CIMED Life Sciences. Pharmalectin and CIMED had no role in data collection, analysis or interpretation of the results.
Conflict of Interest
The corresponding author was the listed Principal Investigator for the trial at Narayana Hrudayalaya Limited and is the Group Head of Clinical Research. His department has received remuneration from the company – Pharmalectin Inc, towards conducting the clinical trials at the center.
David Platt is the CEO of Pharmalectin Inc. However he or his team did not have any access to the data, during the trial and after completion. They did not involve in the statistical analysis and only contributed as a co-author in this manuscript.
REFERENCES
- Team W. Weekly update on COVID-19. In. Emergency Situation Updates. World Health Organization 2020.
- Sanders JM, Monogue ML, Jodlowski TZ, Cutrell JB. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): A review. Jama. 2020;323(18):1824-1836.
- Wevers BA, van der Hoek L. Recently discovered human coronaviruses. Clin Lab Med. 2009;29(4):715-724.
- Gallagher TM, Buchmeier MJ. Coronavirus spike proteins in viral entry and pathogenesis. Virology. 2001;279(2):371-4.
- Huang Y, Yang C, Xu XF, Xu W, Liu SW. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacologica Sinica. 2020;41(9):1141-1149.
- Bertram S, Dijkman R, Habjan M, Heurich A, Gierer S, Glowacka I, et al. TMPRSS2 activates the human coronavirus 229E for cathepsin-independent host cell entry and is expressed in viral target cells in the respiratory epithelium. J Virol. 2013 Jun 1;87(11):6150-6160.
- Fantini J, Di Scala C, Chahinian H, Yahi N. Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int J Antimicrob Agents. 2020;55(5):105960.
- Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020.
- Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol.2016;3:237-261.
- Johannes L, Jacob R, Leffler H. Galectins at a glance. J Cell Sci. 2018;131(9).
- Vasta GR. Roles of galectins in infection. Nat Rev Microbiol. 2009;7(6):424-438.
- Miller MC, Zheng Y, Yan J, Zhou Y, Tai G, Mayo KH. Novel polysaccharide binding to the N-terminal tail of galectin-3 is likely modulated by proline isomerization. Glycobiology. 2017;27(11):1038-1051.
- Dong R, Zhang M, Hu Q, Zheng S, Soh A, Zheng Y, Yuan H. Galectin-3 as a novel biomarker for disease diagnosis and a target for therapy. Int J Mol Med. 2018 Feb 1;41(2):599-614.
- Rodriguez FJ, Scheithauer BW, Roncaroli F, Silva AI, Kovacs K, Brat DJ, et al. Galectin-3 expression is ubiquitous in tumors of the sellar region, nervous system, and mimics: An immunohistochemical and RT-PCR study. Am J Surg Pathol.2008;32(9):1344-1352.
- La Scola B, Le Bideau M, Andreani J, Hoang VT, Grimaldier C, Colson P, et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39(6):1059.
- Jefferson T, Spencer E, Brassey J, Heneghan C. Viral cultures for COVID-19 infectivity assessment: Systematic review. medRxiv. 2020.
- Colton H, Ankcorn M, Yavuz M, Tovey L, Cope A, Raza M, et al. Improved sensitivity using a dual target, E and RdRp assay for the diagnosis of SARS-CoV-2 infection: Experience at a large NHS Foundation Trust in the UK. J Infect. 2020.
- Falzone L, Musso N, Gattuso G, Bongiorno D, Palermo CI, Scalia G, et al. Sensitivity assessment of droplet digital PCR for SARS-CoV-2 detection. Int J Mol Med. 2020;46(3):957-964.
- Caniglia JL, Guda MR, Asuthkar S, Tsung AJ, Velpula KK. A potential role for Galectin-3 inhibitors in the treatment of COVID-19. PeerJ. 2020;8:e9392.
- Arons MM, Hatfield KM, Reddy SC, Kimball A, James A, Jacobs JR, et al. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N Engl J Med. 2020.
- Gandhi M, Yokoe DS, Havlir DV. Asymptomatic Transmission, the Achilles' Heel of Current Strategies to Control Covid-19. N Engl J Med. 2020;382(22):2158-2160.
- Singanayagam A, Patel M, Charlett A, Bernal JL, Saliba V, Ellis J, et al. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Eurosurveillance. 2020;25(32):2001483.
- Harris PA, Taylor R, Minor BL, Elliott V, Fernandez M, O'Neal L, et al. The REDCap consortium: Building an international community of software platform partners. J Biomed Inform. 2019;95:103208.
- Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap): A metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42(2):377-381.
- Eldridge SM, Chan CL, Campbell MJ, Bond CM, Hopewell S, Thabane L, et al. CONSORT 2010 statement: Extension to randomised pilot and feasibility trials. BMJ. 2016;355:i5239.
- Bar-On YM, Flamholz A, Phillips R, Milo R. Science forum: SARS-CoV-2 (COVID-19) by the numbers. Elife. 2020;9:e57309.
- Edros RZ, McDonnell S, Al-Rubeai M. Using molecular markers to characterize productivity in Chinese hamster ovary cell lines. PLOS One. 2013;8(10):e75935.
- Garnett E, Jung J, Tam E, Rajapakshe D, Cheney S, Brown C, et al. Clinical validation and performance evaluation of the automated Vitros Total Anti-SARS-CoV-2 Antibodies assay for screening of serostatus in COVID-19. medRxiv. 2020.
- Salim DK, Telli TA, Tatlı AM, Kılıçkap S, Yumuk PF. Comparison of two different carboplatin and weekly paclitaxel schedule in elderly advanced non-small cell lung cancer patients. Cancer Chemother Pharmacol. 2019 Jun 1;83(6):1137-1145.
- Wang Y, Chang Z, Ouyang J, Wei H, Yang R, Chao Y, et al. Profiles of IgG antibodies to nucleocapsid and spike proteins of the SARS-associated coronavirus in SARS patients. DNA and Cell Biology. 2005;24(8):521-527.
Citation: Sigamani A, Ruthra M, Sudhishma, Shetty S, Madhavi, Chugani A, et al. (2020) Galectin Antagonist use in Mild Cases of SARS-CoV-2:Pilot Feasibility Randomised, Open Label, Controlled Trial. J Vaccines Vaccin. S10:003.
Copyright: © 2020 Sigamani A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.