Indexed In
- Academic Journals Database
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- China National Knowledge Infrastructure (CNKI)
- Scimago
- Ulrich's Periodicals Directory
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- MIAR
- University Grants Commission
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2020) Volume 11, Issue 2
Evaluating the Vaccine Potential of a Tetravalent Fusion Protein against Coronavirus (COVID-19)
Mostafa Norizadeh Tazehkand1* and Orkideh Hajipour22Department of Molecular Biology, Pamukkale University, Denizli, Turkey
Received: 05-Mar-2020 Published: 28-Mar-2020, DOI: 10.35248/2157-7560.20.11.412
Abstract
Coronaviruses are a type of viruses which cause illness ranging from the common cold to other diseases. SARS-CoV-2 is one coronaviruses family that cause respiratory syndrome. The virus first isolated from three people in Wuhan. This virus became known as COVID-19. Common signs of infection comprising of fever, respiratory symptoms, cough, shortness of breath and breathing difficulties. In more severe cases, infection can cause pneumonia, severe acute respiratory syndrome, kidney failure and even death. There is not any vaccine for COVID-19. This study was aimed to design and analysis of recombinant cavvine against COVID-19.
In this research the completely sequence of Envelope and Nucleocapsid protein was fused to multi epitopes (B and MHC I epitopes) obtained from Spike protein and RNA-dependent RNA polymerase and constructed a fusion vaccine.
The vaccine has 621 amino acids which 51 negatively charged residues and 118 positive charged amino acids with 71.906 kDa. The estimated half-life of peptide was found to be greater than 30 hours in mammalian reticulocytes, greater than 20 hours in yeast cells, and greater than 10 hours in E.coli. The instability index II is computed to be 34.81. So, this classifies the protein as stable. The aliphatic index of COVID-19 is found to be 66.86, so the vaccine is probable to be thermostable. The results obtained from protparam and pepcalc analysis revealed that the recombinant antigen is soluble in water. Ramachandran analysis of recombinant antigen showed that 84.3% of amino acids are in most favored regions; this result supported the high-quality structure of the refined model of recombinant vaccine. The result of docking analysis proved that the vaccine has most affinity to HLA B2705-KK10, HLAB3508, HLA-A0201, and HLA B5701. The result of this research revealed that the vaccine has antigenic property and stable structure. The vaccine could be produced by Recombinant DNA technology and expressed in host cells and need to experiment on laboratory animals.
Keywords
COVID-19; Coronavirus; Recombinant vaccine; B-cell
Introduction
Coronaviruses are a type of viruses which cause illness ranging from the common cold to other diseases, for example MERS (Middle East Respiratory Syndrome) and SARS (Severe Acute Respiratory Syndrome). SARS-CoV-2 is one coronaviruses family that cause respiratory syndrome. The virus first isolated from three people in Wuhan. This virus became known as COVID-19 (WHO, 2019).
The COVID-19 is the cause of the 2019-2020 coronavirus epidemic [1].The coronavirus appeared in Wuhan and is believed to have jumped to humans at seafood and animal market where many of the first people to become infected worked (CDC, 2019). The main mode of transmission of COVID-19 is from human to human by respiratory droplets that people respire during sneezing and coughing [2]. According to the Centers for Disease Control and Prevention (CDC), the coronavirus' incubation period is believed to be 2 to 14 days. However, one case is reported as having an incubation period of 27 days [3].
Common signs of infection comprising of fever, respiratory symptoms, cough, shortness of breath and breathing difficulties. In more severe cases, infection can cause pneumonia, severe acute respiratory syndrome, kidney failure and even death [1]. No drug has yet been discovered to treat COVID-19 infections. Antivirals being tested include chloroquine, darunavir, galidesivir, interferon beta, the lopinavir/ritonavir combination, the RNA polymerase inhibitor remdesivir, and triazavirin [4,5]. Remdesivir and chloroquine effectively inhibit the coronavirus in vitro [6]. There is not any vaccine for COVID-19. The United States NIH (National Institutes of Health) is collaborating with Modern to generate an RNA vaccine from spike protein of the coronavirus. In another research, Inovio Pharmaceuticals is developing a DNA vaccine [7]. The University of Queensland (Australia) is studying the potential of a molecular clamp vaccine that would genetically modify viral proteins in order to stimulate an immune reaction [8]. Another research is doing by Canadian scientist In Canada (University of Saskatchewan), the scientists are working on a vaccine aiming to start animal testing in March 2020 and human testing in 2021 [9]. Preliminary results from a metacentric trial, announced in a press conference and described by Gao, Tian, and Yang, suggested that chloroquine is effective and safe in treating COVID-19 associated pneumonia, improving lung imaging findings, promoting a virus-negative conversion, and shortening the disease course [2].
Envelope protein, Spike glycoprotein, RNA-dependent RNA polymerase, and Nucleocapsid Protein have significant roles in pathogenesis of COVID-19. So, in this study, the epitopes of Spike protein and RNA-dependent RNA polymerase fused to Envelope protein and Nucleocapsid Protein to design a peptide vaccine against COVID-19. Then, the vaccine was analyzed with different bioinformatics analysis and software ’ s. After that molecular docking analysis was used to recognize the affinity of vaccine to different MHC-I.
Materials and Methods
In this research the completely sequence of Envelope and Nucleocapsid protein was fused to multi epitopes obtained from Spike protein and RNA-dependent RNA polymerase and constructed a vaccine.
Firstly, the antigenicity, allergen city and toxicity selected proteins were analyzed by vaxijen, Algpred and Toxinpred [10,11]. The result of those analysis showed that the proteins have high antigenic score and are not toxic. B-cell epitopes were predicted by Immune Epitope Database server which is a free webserver for designing of bacterial and virus vaccines. In our study the epitopes higher than 0.35 thresholds were selected to B-cell epitopes [12].
IEDB and PropredI were used to prediction of MHC class I epitopes. In this research the epitopes were evaluated for their binding affinity with different HLA alleles (p values<0.05 were considered significant) [13].
In this study we used from KK (Lysine-Lysine) linker for linking of B-cell and T-cell epitopes. The antigenicity, allergen city, and toxicity of fusion protein were tested by vaxijen, Algpred, and Toxinpred software ’ s [10]. The toxic sequence of designed protein was removed from vaccine structure. Afterward, molecular weight, half-life, aliphatic index, isoelectric point instability index, and stability of COVID-19 vaccine were tested by Papcolc and Protparam webserver [14].
We used from Parabi webserver for prediction of Trans membrane helices of vaccine. 3D structure of protein was drowning by SWISS-MODEL. The SWISS-MODEL webserver is a computerized modeling program which develops a protein 3D structure model of an unknown structure protein based on the sequence similarity with the known structured protein [15]. For docking analysis, we need a refine model of proteins, for this reason the 3D structure of vaccine was refined by 3D refine analyzing software. 3Drefine is software for computationally efficient protein structure refinement with the ability to accomplish web based visual and statistical investigation. The 3Drefine software uses iterative optimization of hydrogen bonding network combined with atomic-level energy minimization on the optimized model using a composite physics and knowledge-based force fields for efficient protein structure refinement. The software exposed five refıned model to our vaccine. The refined models were patterned for 3D refine score, RMSD score, GTD-TS, GDT-HA score and MolProbity score. The best model was taken and the chosen model was examined via Procheck Ramachandran plot analysis [16].
Docking analysis of recombinant vaccine
Protein-protein docking of recombinant vaccine was done by HEX protein protein docking analysis by considering HLA B2705-KK10, HLAB3508, HLA-A*02, and HLA B5701(the types of MCH class I) as a receptor and designed vaccine as a ligand. For this reason, molecular structure of different MCH-I was taken in PDB format from Protein Data Bank (PDB). Hex (http://hexserver.loria.fr/) is the first Fourier transforms based protein docking software to be powered using graphics processors. The software requires the structure of proteins in PDB format to be uploaded and start it produces a ranked list of up to 1000 docking predictions. In our research 3D structure of HLA B2705-KK10, HLAB3508, HLA-A0201, and HLA B5701 are used as receptors and our designed vaccine structure was used as a ligand were uploaded to Hex protein webserver [17].
Results and Discussion
The protein sequence of Spike protein and RNA-dependent RNA polymerase fused to Envelope protein and Nucleocapsid Protein were obtained from NCBI. The completely sequence of Envelope and Nucleocapsid protein was fused to multi epitopes obtained from Spike protein and RNA-dependent RNA polymerase and constructed a vaccine.
The antigenicity of these sequences was tested by VaxiJen server. The vixen score of these sequences were 0.6298 (Envelope protein), 0.4646 (Spike protein), 0.4064 (RNA-dependent RNA polymerase), and 0.5522 (Nucleocapsid Protein) respectively. The score higher than threshold (0.4) have good antigenicity; therefore all proteins were suitable for our research. B cell epitopes from Spike protein and RNA-dependent RNA polymerase were predicted by Immune Epitope Database and Analysis Resource server. The B cell epitopes that having higher than 0.35 were chosen to further analysis. The MHC class I epitopes from Spike protein and RNA-dependent RNA polymerase were predicted by vaxign server. The epitopes were assessed for their binding affinity with predominant HLA I alleles (P-values < 0.05). The selected epitopes fused to Envelop and Nucleocapsid protein by KK linkage. The sequence of our designed vaccine with 621 amino acids is shown in Figure 1.
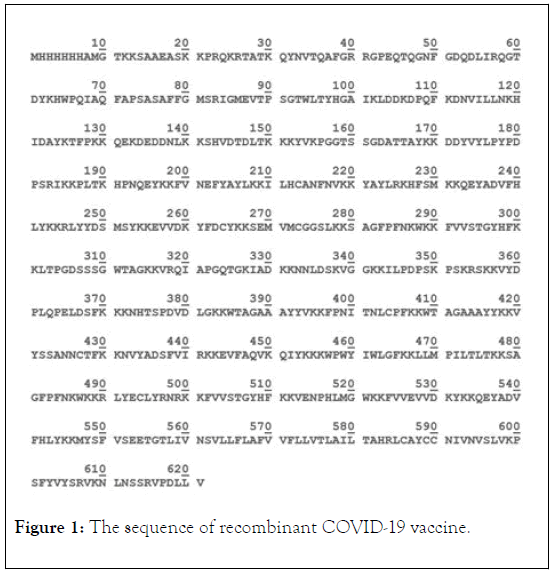
Figure 1: The sequence of recombinant COVID-19 vaccine.
The antigenicity, allergenecity, and toxicity of fusion vaccine by analyze different software’s. The result of analysis showed that the vaccine has high antigenic score and does not allergic or toxically effect on human cells (Figures 2-4).
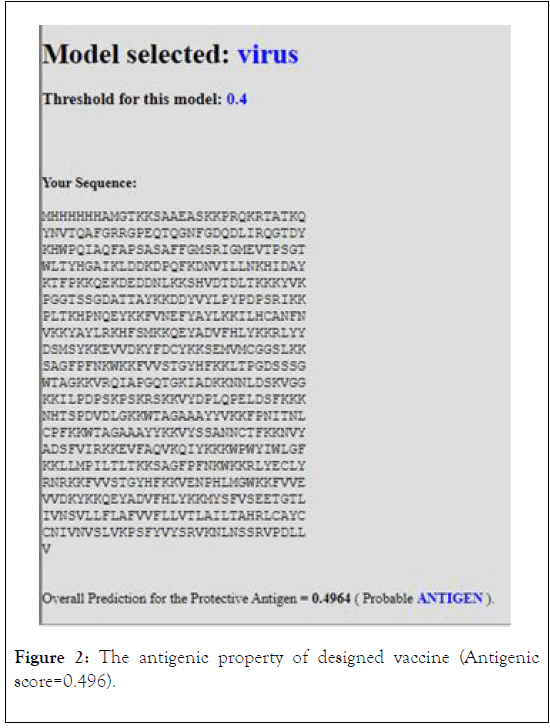
Figure 2: The antigenic property of designed vaccine (Antigenic score=0.496).
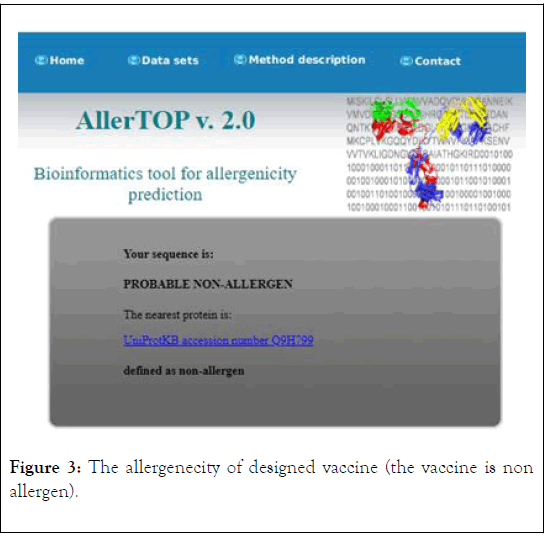
Figure 3: The allergenecity of designed vaccine (the vaccine is non allergen).
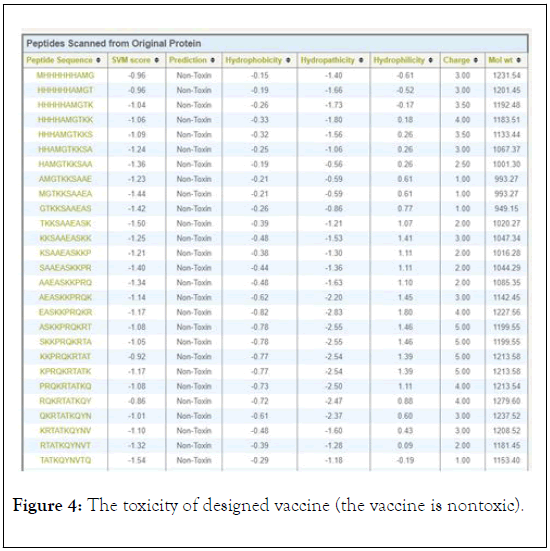
Figure 4: The toxicity of designed vaccine (the vaccine is nontoxic).
The vaccine has 621 amino acids which 51 negatively charged residues and 118 positive charged amino acids. The physicochemical property of COVID-19 vaccine was analyzed via protparam and the result showed that molecular weight of candidate vaccine is 71.906 kDa. The estimated half-life of peptide was found to be greater than 30 hours in mammalian reticulocytes, greater than 20 hours in yeast cells, and greater than 10 hours in E.coli. The chemical formula of recombinant vaccine is C3315H5113N871O881S20 with 10200 atoms.
The instability index II is computed to be 34.81. So, this classifies the protein as stable. The grand average of hydropathicity of designed antigen is -0.637, thus the vaccine is a hydrophilic protein and likely interact with water. The aliphatic index of COVID-19 is found to be 66.86, so the vaccine is probable to be thermostable. The results obtained from protparam and pepcalc analysis revealed that the recombinant antigen is soluble in water. The result from Parabi showed that the membrane helices value of vaccine was 35.27% (Figure 5). The vaccine does not have any Tran’s membrane helix, so the vaccine simply produced and no expression difficulties are predicted in the expression of antigen.
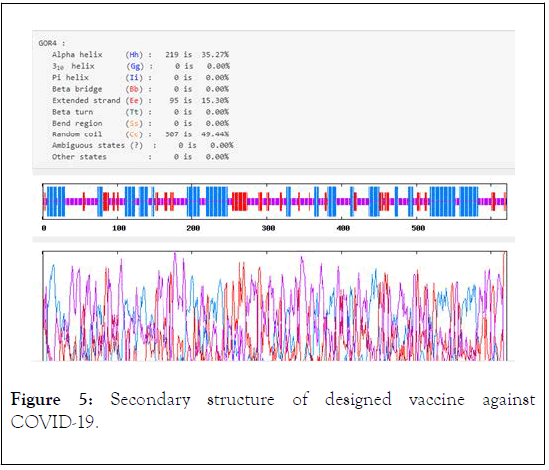
Figure 5: Secondary structure of designed vaccine against COVID-19.
In this study we used from SWISS NODEL software to drawing of 3D structure of Coronavirus vaccine and then, the vaccine structure was refined by 3Drefine. The result of 3D refine analysis showed that 3Drefine score is 10167, GTD-TS score is 1.000, GTD-HA score is 0.9964, RMSD score is 0.219, MolProbity score is 2.093, and RW Plus score is -19505 (Figure 6). The selected model from 3drefine was examined by Ramachandran plot analysis using Procheck webserver.
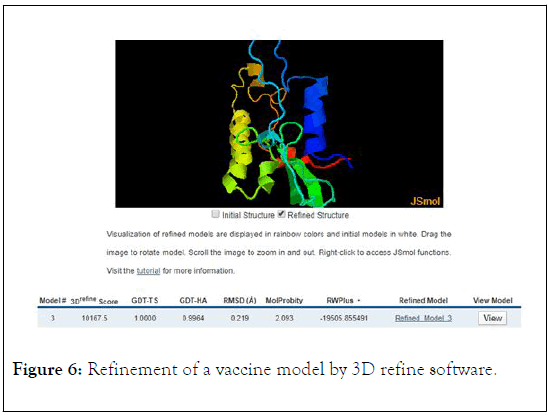
Figure 6: Refinement of a vaccine model by 3D refine software.
Ramachandran analysis of recombinant antigen showed that 84.3% of amino acids are in most favored regions, 10.2% of residues are additional allowed regions, 2.4% of amino acids in generously allowed regions, and just 3.1% of amino acids are in disallowed regions (Figure 7). This result supported the highquality structure of the refined model of recombinant vaccine.
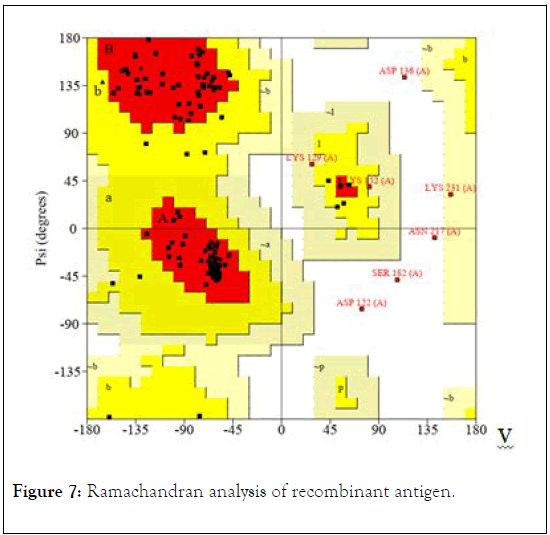
Figure 7: Ramachandran analysis of recombinant antigen.
The Pepcalc analysis showed that the vaccine has the net charge of vaccine at pH 7 is 68/3 and the isoelectric point of vaccine is 10.32 (Figure 8).
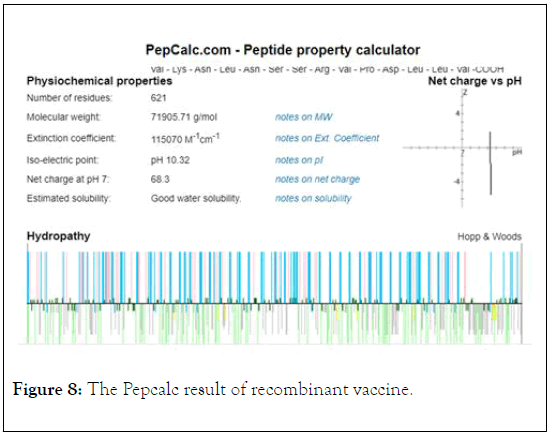
Figure 8: The Pepcalc result of recombinant vaccine.
Docking analysis of recombinant vaccine
Protein-protein docking of recombinant vaccine was done in HEX protein docking analysis by considering HLA B2705-KK10, HLAB3508, HLA-A0201, and HLA B5701 (the types of MCH class I) as a receptor and designed vaccine as a ligand. The result of protein docking analysis showed that maximum affinity of recombinant vaccine to HLA B2705-KK10, HLAB3508, HLAA* 02, and HLA B5701 with the score of -716/75, -691.28, -693.60, and -557.87 respectively (Figure 9).
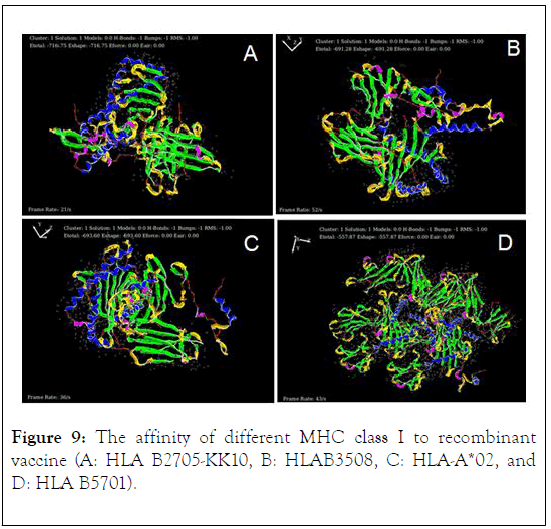
Figure 9: The affinity of different MHC class I to recombinant vaccine (A: HLA B2705-KK10, B: HLAB3508, C: HLA-A*02, and D: HLA B5701).
In the present study the selected proteins have the antigenic score higher than threshold (0.4) which can be candidate to effective vaccine against COVID-19. Having above threshold scores mean that the vaccine can be recognized by MHC-1 and B-cells. Physicochemical property of designed vaccine against COVID-19 revealed that the antigen had a molecular weight of 71 kDa. The researchers showed that the proteins with high molecular weight are more stable than low molecular weight proteins [18]. The half-life estimated of recombinant vaccine is greater than 30 hours in mammalian cells, higher than 20 hours in yeast cells, and higher than 10 hours in E.coli. The result revealed that the recombinant vaccine can be overexpress in E.coli or other host cells. Beside that the produced vaccine can be used in human, because the vaccine can be stable 30h in human cells [19-21]. The Instability index score of recombinant vaccine is 34.81 which lower than 40, the score revealed that the vaccine is considered as stable. Our designed vaccine does not have allergenecity and toxicity effect on human and animal cells. The stability and water solubility of peptide vaccine is important and our designed vaccine is stable and soluble in water. Thus the recombinant vaccine is not estimated to drop harmful allergic responses in humans. The result of docking analysis proved that the vaccine has most affinity to HLA B2705-KK10, HLAB3508, HLA-A0201, and HLA B5701.
Conclusion
The result of our research showed that the vaccine can activate cellular and humoral immune responses against COVID-19. The vaccine had suitable structural, physiochemical, and immunological properties. Nevertheless, the vaccine could be produced by Recombinant DNA technology and expressed in host cells and need to experiment on laboratory animals.
REFERENCES
- Hui DS, I Azhar E, Madani TA, Ntoumi F, Kock R, Dar O, et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health: The latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis. 2020; 91: 264-266.
- Nebehay S, Kelland K, Liu R. WHO: No known effective' treatments for new coronavirus 2020.
- Ebehay S, Kelland K, Liu R. WHO: No known effective' treatments for new coronavirus. Thomson Reuters 2020.
- Chen H, Guo J, Wang C, Luo F, Yu X, Zhang W, et al. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: A retrospective review of medical records. The Lancet. 2020
- Li G, De Clercq E. Therapeutic options for the (2019-nCoV) novel coronavirus 2019.
- Mishra M. Johnson & Johnson working on vaccine for deadly coronavirus. 2020.
- Steenhuysen J, Kelland K. With Wuhan virus genetic code in hand, scientists begin work on a vaccine. 2020.
- Chung M, Bernheim A, Mei X, Zhang N, Huang M, Zeng X, et al. CT imaging features of 2019 novel coronavirus (2019-nCoV). Radiology. 2020; 4: 200-230
- Duddu P. Coronavirus outbreak: Vaccines/drugs in the pipeline for Covid-19 at the Way back Machine Archived. 2020.
- Doytchinova IA, Flower DR. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007; 81: 4.
- Golshani M, Oloomi M, Bouzari S. In silico analysis of Shiga toxins (Stxs) to identify new potential vaccine targets for Shiga toxin-producing Escherichia coli. In silico pharmacol. 2017; 5: 2.
- Srivastava S, Kamthania M, Singh S, Saxena AK, Sharma N. Structural basis of development of multi-epitope vaccine against middle east respiratory syndrome using in silico approach. Infect Drug Resist. 2018; 11: 2377.
- Dikhit MR, Kumar A, Das S, Dehury B, Rout AK, Jamal F, Sahoo GC, et al. Identification of potential MHC Class-II-restricted epitopes derived from Leishmania donovani antigens by reverse vaccinology and evaluation of their CD4+ T-cell responsiveness against visceral leishmaniasis. Front Immunol. 2017; 8: 1763
- Pritam M, Singh G, Swaroop S, Singh AK, Singh SP. Exploitation of reverse vaccinology and immunoinformatics as promising platform for genome-wide screening of new effective vaccine candidates against Plasmodium falciparum. BMC Bioinform. 2019; 19: 468.
- Ghasemi A, Ranjbar R, Amani J. In silico analysis of chimeric TF, Omp31 and BP26 fragments of Brucella melitensis for development of a multi subunit vaccine candidate. Iran J Basic Med Sci. 2014; 17: 172.
- Bhattacharya D, Nowotny J, Cao R, Cheng J. 3Drefine: An interactive web server for efficient protein structure refinement. Nucleic Acids Res. 2016; 44: 406-409.
- Norizadeh Tazehkand M, Hajipour O. In silico Design a vaccine Candidate against Corynebacterium diphtheriae. Int J Mole Clin Microb. 2019; 9: 1082-1089.
- Naz K, Naz A, Ashraf ST, Rizwan M, Ahmad J, Baumbach J, et al. PanRV: Pangenome-reverse vaccinology approach for identifications of potential vaccine candidates in microbial pangenome. BMC Bioinform. 2019; 20 :123.
- Dhama K, Sharun K, Tiwari R, Sircar S, Bhat S, Malik YS, et al. Coronavirus Disease COVID 2019.
- Centers for Disease Control and Prevention. Coronavirus Disease 2019 (COVID-19) 11 February 2020. Archived from the original on 23 February 2020.
- Ghoorah AW, Devignes MD, Smaïl-Tabbone M, Ritchie DW. Protein docking using case-based reasoning. Proteins: Structure, Function, and Bioinform. 2013; 81: 2150-2158.
- Norizadehtazehkand M, Hajipour O. Multi Epitope Vaccine Candidate against Mycobacterium Tuberculosis. Drug Des Int Prop Int. 2019; 3: 351-358.
Citation: Tazehkand MN, Hajipour O (2020) Evaluating the Vaccine Potential of a Tetravalent Fusion Protein against Coronavirus. J Vaccines Vaccin. 11:412. DOI: 10.35248/2157-7560.20.11.412
Copyright: © 2020 Tazehkand MN, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

