Indexed In
- RefSeek
- Hamdard University
- EBSCO A-Z
- Publons
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Mini Review - (2021) Volume 6, Issue 2
Emergence of New Drug Discovery Paradigm using Computational and Experimental Biology: Case Study of using Repurposing Drugs for COVID-19
Sreedhara Voleti1*, Shalini Saxena1 and Uday Saxena22Reagene Innovations Private Limited, School of Life Sciences, University of Hyderabad, Hyderabad, Telangana, 500046, India
Received: 01-Jul-2021 Published: 22-Jul-2021, DOI: 10.37421/2576-389X.2021.6.152
Abstract
Discovery of new drugs is an expensive and time-consuming process. Average time taken from bench to bedside (idea to drug launch) is about 12 years including clinical trials. The emergence of COVID-19 pandemic has demonstrated that there is a dire need to be able to launch therapeutics much sooner than the traditional timelines. As such the industry is reinventing the process to deliver drugs much faster.
As such we present here a new paradigm of identifying drugs for COVID-19. It uses three fundamental principles, viz., 1) using existing FDA approved drugs which have been approved for other diseases, so their safety is proven, 2) Using computational rational approaches for choosing the best drugs against the target of interest such as the Receptor Binding Domain of the Spike Protein of COVID-19, and, finally, 3) validation of these short-listed drugs through in vitro disease models. We estimate that such an approach could crash the timelines of bench to bedside to 5 years or less, thus fulfilling the needs of urgent unmet diseases such as COVID-19 faster.
Keywords
In-silico; in-vitro disease models; COVID-19; Computational biology; Drug discovery
Introduction
Design is the key of life! Design brings a progressive, tractable, and visual path for any walk of life let it be agriculture, architecture, engineering, healthcare, and by and large encompassing sciences of various flavors such as biology, chemistry, pharmacology, and toxicology etc. Design in human life since centuries brought marvelous outcomes human can enjoy, while till late 19th century, it was limited by the art of human hands. Ever since the advent of computers the architecture of computer applications through programming and visual aids growing research has encompassed, infused, and now, made it inseparable along the multidisciplinary fields – where the conventional healthcare has now demanding its use. From the late 1970’s, the spread of computers in various science and healthcare branches made scientists not just use, but made it a way of life, have termed it “in silico” [1-4]. Now there are innumerable journals specialized publishing research with in silico approaches, text books filled with chapters on in silico theories and applications, even patents on in silico outcomes. The in silico theories, processes, methods, and approaches led to researchers deriving properties brought in “informatics” as broad nomenclature which forms the basis/key for particular field to make one to use design principles through computers to formulate future research science endeavors on practical applications.
Healthcare of living beings is such a field that, for centuries, used applications of agents (natural or synthetic) in definitive proportions and formulations leading to a healthy life, that every human long for. Chemical, biological, pharmacological, toxicological and even clinical nature–handled by experimentalists and physicians were the standard process of finding a right agent/s for a right disease for living beings, called as a Therapeutic, or simply “Drug”. Discussed vide infra, the process of identification of a drug, a path called “drug discovery” is no longer archaically followed by experimentalists, but they now heavily rely on computational tools which help reduce the companies the burden of costs, time, and speed, but more importantly, rationalize the process of drug discovery–leading to faster outcomes with clear increased chance and enhanced probability of success of finding a drug.
Pharmaceutical industry (especially small molecule focused therapeutics) have made millions of molecules in understanding the so called structure-activity/function/toxicity-relationships (SAR/SFR/STR) and almost more number of experiments (at least 3 times larger) were annotated for their biological, pharmacological, and toxicological portfolio. Ever since computational tools coupled with statistical and visual data analysis came into existence since 80s, the landscape of drug discovery changes, and evolved was the Lipinski’s Rule-of-5 (RO5) [5,6] and many variants of such for oral administration drugs and other forms of drug evaluations. Subsequently, use of in silico technologies have further been implemented significantly for knowledge-generation, pattern recognition, and funnel-down evaluation-thus, enabling the “virtual” experiments prior to “real” experiments prioritizing the odds of success. This process has multifold advantage by bringing down the resource optimization, costs, time, and more importantly the rationale for doing an experiment that gives out an optimized start-up. This process thus led to tossing the term “Drug Design”, without which a small-molecule drug discovery wouldn’t start. Thus, the in silico approaches such as structure-based, ligand-based, and virtual screening methods have an in separable inclusion in the process of hit identification to lead optimization, while many pharmaceutical industries of novel drug design/discovery R&D practice have infused incorporation of computational tools a priori, for a rational start. Following are some of the techniques used largely in computational tools.
In silico methodologies are playing an important role for the identification of promising drug candidates in the drug discovery process. Drug research and development starts with the identification of the bimolecular targets for an intended treatment and proceeds with the high-throughput screening experiments to identify bioactive compounds for the defined targets, together with the corresponding bioactivity levels. There are various in silico methods listed in Figure 1 that were used in the drug discovery process with generic examples such protein- ligand interactions and target-based discovery from structure-based methods. The QSAR and Pharmacophore or feature- based methods in ligand-based drug design.
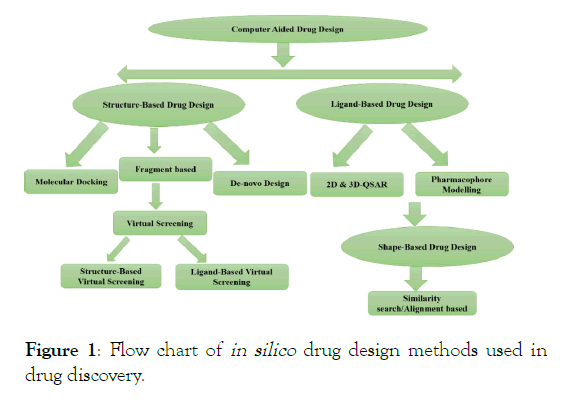
Figure 1: Flow chart of in silico drug design methods used in drug discovery.
Structure-Based Drug Design
Structure-based drug design executed with available structural models of the target proteins, which are provided by X-ray diffraction, nuclear magnetic resonance (NMR) or molecular simulation (homology modelling of protein). The crystal structure of a ligand bound to a protein provides a detailed insight into the interactions made between the protein and the ligand [7]. Designed structure can be used to identify the sites where the compound can be changed to modulate the physicochemical and ADME properties of the compound. It can also show which parts of the compound are important to affinity and which parts can be altered without affecting the binding. With this provided information molecular database is searched against the target protein to identify the suitable molecules whose molecular shapes match the binding sites of the receptor and binding affinity is high. Then, these molecules are synthesized/procured for their biological activities for further drug development. In short, structure-based drug design plays an extremely important role in drug design. The structure based methods include molecular docking, de novo design, structure-based virtual screening and fragment based discovery.
Homology modeling
An integral part of structure-based design is the construction of a protein structure in 3-dimensions using the primary sequence of the desired target with the templates chosen from the sequence similarity, sequence identity, and structural-homologs either from the sub-types or from the family tree. Several methods such as BLAST [8], are employed for sequence comparison, alignment, and further construction of back-bone in Homology modelling. Once an approximate structure of the target protein is obtained, it is further subjected to energy minimization to get to a local minimum, and further subjected to molecular dynamics studies for getting an average of ensemble structure that could be closer to the equilibrium structure.
Molecular dynamics (MD) simulations
Molecular Dynamics gives insights into the stability and dynamic properties of the proteins and Protein-Ligand complexes. MD simulations provide detailed insight of protein stability over an equilibrated time-length (such as nano or micro seconds), and subsequent averaged protein-ligand interactions in motion contributing for their stable bound conformation [9]. The outcome is to visualize the effect of ligand binding on protein conformational changes, intra- and inter-molecular interactions stabilizing the complexes. Molecular simulations can be performed under conditions where it is difficult or impossible to conduct experiments-for instance, at very high pressures and temperatures. In drug discovery and molecular science applications molecular dynamics have made important developments in advancing drug discovery. MD, especially when coupled with these other computational tools, will open the door to addressing the many drug discovery problems for which the dynamic nature of proteins cannot be ignored, as in the mechanisms of highly mobile membrane proteins and in ligand- induced conformational changes of active sites.
Molecular docking
Molecular docking is a useful tool used to predict the geometry and to score the interaction of a protein in complex with a small-molecule ligand. Docking can be used to predict if a given drug is potentially able to bind to the target protein. Docking studies have been successfully exploited in drug repurposing approach [10]. Molecular docking is in fact a convenient and fast method to screen large libraries of both ligands and targets, with a full range of sampling options, and its restricted to studies where a 3D structure of the target is available through crystallography, NMR, or homology models. The information obtained from the docking technique can be used to suggest the binding energy, free energy and stability of complexes. The equilibrium between target protein and ligand is directed by the free energy of the complex compared to the free energy of the individual target and ligand. This includes not only the interaction between target and ligand but also the solvation and entropy of the three different species and the energy of the conformation of the free species. The main objective of molecular docking is to achieve ligand-receptor complex with optimized conformation and with the intention of possessing less binding free energy.
Virtual Screening (VS)
It plays an important role within the pharmaceutical industry in lead discovery process. VS refers to computational screening of large libraries of chemicals for compounds that complement targets of known structure which could be tested experimentally. VS can be performed either by docking a known drug into a large set of different target structures, or by docking a database of approved drugs into one intended specific target. Since, it takes place in the three-dimensional active site of the target, it is also called as structure-based virtual screening.
Denovo design
It attempts to use the unliganded structure of the protein to generate novel chemical structure that can bind to the protein. There are varying algorithms, most of which depend on identifying initial putative sites of interaction that are grown into complete ligands.
Fragment-based drug discovery
It is based on the premise that most ligands that bind strongly to a protein active site can be considered as a number of smaller fragments or functionalities. Fragments are small molecules that are identified by screening a relatively large number of libraries of molecules (in the range of 20,000) with molecular weight in the range of 200-300 by X-ray crystallography and NMR spectroscopy. These fragments binding to a given protein can further be used to design new final ligands by adding appropriate linkers between the fragments or by incorporating features of the fragment onto existing ligands [11]. Several pharmaceutical companies have adapted fragment-based drug discovery in enhancing the potency and pharmaceutically acceptable features such as Rule-of-5 and other physiochemical features.
Ligand based drug design
In ligand-based drug design, structural features of known compounds are used to construct in silico models that are used to predict the properties of other compounds. Ligand-based approaches are based on the concept that similar compounds tend to have similar biological properties. This concept of similarity has been extensively utilized in lead discovery and optimization primarily because it takes into account the poly pharmacological nature of drugs [12]. These other molecules may be used to derive a pharmacophore model that defines the minimum necessary structural characteristics a molecule must possess in order to bind to the target. In other words, a model of the biological target may be built based on the knowledge of what binds to it and this model in turn may be used to design new molecular entities that interact with the target. The 2D and 3D-QSARs, molecular similarity search and pharmacophore modelling are the ligand-based design techniques.
Structure Activity Relationships (SAR)
In the absence of a protein-structure and where a protein- structure cannot be generated using Homology Modeling, SAR studies will bring mathematical and statistical correlation relationship between various physiochemical parameters of small molecules to their biological/pharmacological properties when interact with the macromolecules in multi-dimensions. Any correlation above 3-dimensions will lead to quantitative relationship, and within one- and two-dimensions, they bring qualitative correlations. It contains a correlation between calculated properties of small molecules such as MWt, LogP, and LogD etc. to their experimentally determined biological activity of a target, or properties such as absorption, distribution, and metabolism. Quantitative SAR is mostly applied to pharmacological activities such as IC50, Ki, or LD50, while qualitative SAR is applied to metabolism, relative rates of absorption etc. Drug discovery often involves the use of QSAR to identify chemical structures that could have good inhibitory potential on specific target. Since 1990s, 3D structural information was introduced into the QSAR methods, namely 3D-QSAR with the improvement of computing power, conformational analysis, and co-crystal structure information for initial geometry of the molecules. It is a powerful tool for studying the interactions between drugs and target proteins with establishing the relationship of drug structure activity relationship.
Pharmacophore modeling
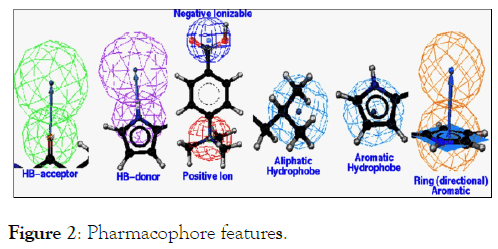
A pharmacophore is an abstract description of molecular features of a given molecule, which are essential for molecular identification and recognition of a ligand by a biological macromolecule. Typical pharmacophoric features include hydrophobic centroids, aromatic rings, hydrogen bond acceptors, hydrogen bond donors, positive charge and negative charge (Figure 2). It depends on atomic properties rather than element types; it does not depend on specific chemical connectivity [13]. A pharmacophore model can be established either in a ligand-based manner, by superposing a set of active molecules and extracting common chemical features that are essential for their bioactivity, or in a structure-based manner, by identifying a common sub-fragment features between the macromolecular targets and ligands. A Pharmacophore map can be generated by superposition of active compounds to identify their common features. The aim of pharmacophore mapping is to transform 2D structure-activity information into the 3D requirements for binding to the target protein.
Figure 2: Pharmacophore features.
Pharmacophore model allows one to search 3 dimensional structure of molecules in databases for alternate (hopping) other molecules that matches.
Shape based screening
Shape-based methods for aligning and scoring ligands have proven to be treasured in the field of CADD. It is a powerful shape-based flexible ligand superposition and virtual screening method, which rapidly produces accurate 3D ligand alignments and proficiently enriches actives in virtual screening process [14]. There are 3 key considerations emphasized when developing Shape Screening:
1. The speed at which structures can be processed.
2. The quality of the superposition.
3. The ability to selectively identify actives over decoys within database.
In silico and in vitro dual synergistic paradigm for rapid and rational identification of therapeutics for COVID-19
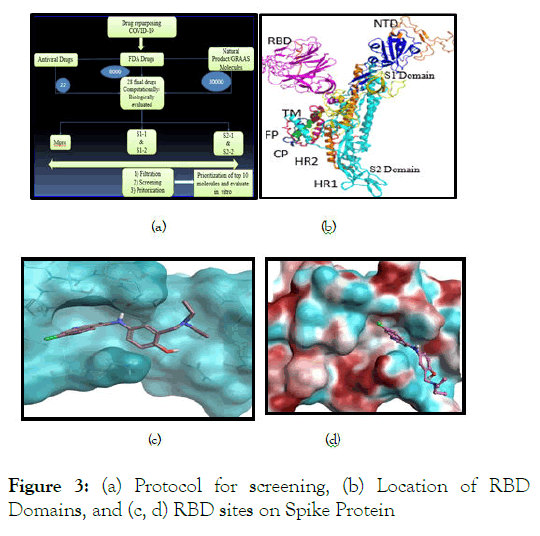
SARS-COV-2 as pandemic has rocked the world for identification of mitigating agents to resolve the human health. Broad options for such relief, proposed by knowledgeable scientists include to either to kill the virus through therapeutics (small molecule, natural product, and herbal nature) or inactivate/protect it from anti-viral or new vaccines. Since time and safety are the key essence in this regard, conventional new drug discovery wouldn’t be of any help let alone, synthesis of large quantities of existing anti-virals for the worldly needs is not possible in such short time without having any rational proof of their utility. Many groups across the globe have solved the structure of the Spike Protein of SARS-COV-2 with the Main Protease was extensively studied with co-crystal information with a peptidomimitic molecule which has greater potential inhibitory activity. The receptor binding domain (RBD) of spike protein also has been identified, but very few reports have done extensive analysis on this part of protein. Inhibiting the RBD domain of the spike protein is one of the thoughts INDRAS (computational) (experimental) came up with idea to explore the entire drug repository and natural products for prioritizing top molecules with rational success, and Reagene Innovations to perform key in vitro experiments to prove the odds of success based on in silico studies. The outcome of such study within one year has yielded few widely referred publications. The in silico-in vitro duet thus yielded Ertugliflozin, a drug originally prescribed for T2DM that controls blood glucose through inhibition of SGLT-2 protein, which demonstrated immense inhibitory potential of RBD of Spike protein of SARS-COV-2 and ACE2 (human) association and many other in vitro studies that are highly relevant (such as secretion of IL-1β, protection of thrombogenic potential, accumulation of monocytes in lungs etc.). The overall in silico process setup is given below (Figure 3) detailing the process and S1/S2 domains of RBD of Spike Protein).
Figure 3: (a) Protocol for screening, (b) Location of RBD Domains, and (c, d) RBD sites on Spike Protein
In vitro experimental validation of computationally prioritized drugs
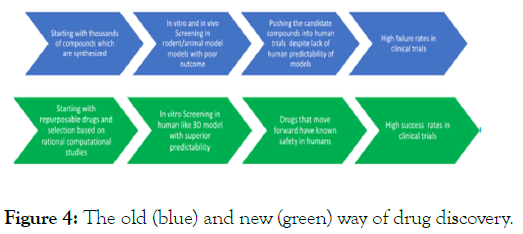
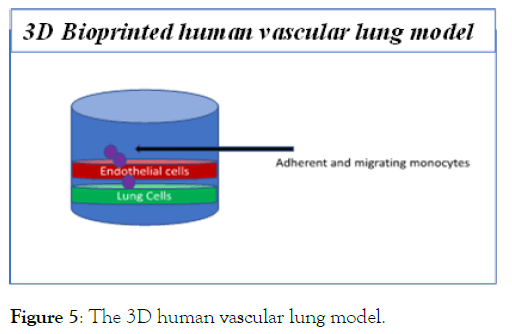
It is mandatory to test the findings of in silico studies in experimental studies, otherwise the results are less meaningful. Drug discovery is not limited to theoretical calculations, therefore backing up the findings of in silico results by experimental methods is mandatory to make go/no go decisions on the viability of drugs. The pharmaceutical industry relies heavily on in vitro followed by animal model and finally clinical testing of drugs before market launch. No drug is approved without such experimental laboratory and clinical studies. Experimental methods are designed to mimic the disease steps that are believed to occur in the human disease pathogenesis. Since we are using COVID-19 as a case study here, we can summarize the different steps of how SarsCoV2 infects humans and causes disease and then design experimental studies to test the drugs. We have designed a human like 3D bioprinted vascular lung model to better mimic human biology and disease. The major problem with current drug discovery efforts is major reliance on animal models. Animal models of disease, especially rodent, do not accurately capture human disease simply because they are so far removed from humans. At best the closest are the non-human primate models, which are expensive and often not feasible. So there exists a huge gap in turns of predicting human out comes by only relying on rodent models. Thus there is a need to find newer human like models to improve drug discovery success rates. Figure 4 shown below is the way typical drug discovery is performed (hit or miss model) versus our proposal of a new paradigm. The 3D human vascular lung model we developed is shown in Figure 5.
Figure 4: The old (blue) and new (green) way of drug discovery.
Figure 5: The 3D human vascular lung model.
The first step in COVID-19 begins with entry of the virus thru nasal and oral passages into the lungs. Once the virus reaches the lungs it uses its surface protein spike protein 1 to attach to lung cells and gain entry. Specifically, as described above, there is a receptor binding domain with in the spike protein that engages with the ACE2 receptor with high affinity. This binding triggers the entry of the virus into the lung cells. Conversely, if this binding is interrupted, then the virus cannot enter the cells. This inhibition of RBD binding to ACE2 becomes an avenue for finding drugs that may be used in treatment of COVID-19 in early stages of infection.
A simple screening assay can be designed where the purified ACE2 receptor protein is coated onto ELISA plates and labelled RBD is added with or without the drug of interest. The binding is monitored by detecting the labelled RBD (usually labelled by biotin in an ELISA system) to the plate and drug inhibition is described as decreased binding of RBD. In our studies published [15-17] we have used this simple technique to corroborate the computational prediction of drugs inhibiting RBD-ACE2 binding and found high degree of correlation. Thus the first step in COVID-19 disease process can be modelled.
The next step is believed to be the uncontrolled production of cytokines by the lung cells as a defensive mechanism to fight the viral infections. Typically, inflammatory cytokines such as IL-1β are involved in this step and results in “cytokine storm” observed in humans. If left unchecked, cytokine storm can create havoc in the lungs and therefore this event has also become a therapeutic target in fighting the disease. The “cytokine storm” can be mimicked in vitro by culturing human endothelial cells and lung cells and measuring IL-1β secretion to simulate this event. We have designed a new 3D bioprinted human vascular lung model. The lung is a heavily vascularized organ and therefore to accurately depict lung functions, it is better to use both endothelial cells as well as lung epithelial cells. The 3D model is a bioprinted layer of endothelium and the lung epithelium. To measure “cytokine storm” media from the 3D layers are collected after drug treatment and IL-1β secretion is measured. Thus this is an elegant system to measure the effect of drugs on “cytokine storm”.
Another related event that is observed in infected patient lungs is the massive influx of circulating monocytes – presumably to try and kill the virus. This event can also be measured using the 3D human vascular lung model. Monocytes can be added to the 3D system and their adhesion, which is the first step in their entry to the lung, can be measured. Drugs that prevent this adhesion would be anti-inflammatory and can be useful in COVID-19.
Another critical observation that’s been seen in COVID-19 patients is thrombosis, or clotting of blood vessels. In fact, blood clotting or endothelitis has now become the centre of attention and is believed to be an important reason for death in COVID-19 patients. We are able to measure this thrombosis in our 3D printed system by using the secretion of a biomarker called thrombomodulin, an anti-coagulant protein which normally resides on endothelial cell surface but is released during inflammation, thus making the endothelial surface less thrombogenic. The mortality rate of COVID-19 in-hospital patients has been directly linked to the increased levels of this biomarker in circulation.
Conclusion
Typical drug discovery is time consuming, expensive with low probability of success. Newer models of discovering drugs have to be invented to serve the patients especially in times such as COVID-19 pandemic. In this mini review, we have proposed combining computational studies along with targeted in vitro studies to rapidly identify drugs that can be repurposed to fight COVID-19. Traditional drug discovery would require a 10 plus years to get to market launch. Our new paradigm of combining screening in silico studies of large numbers of marketed drugs, whittling them down a smaller number and testing them in innovative in vitro human like model that directly mimic COVID-19 in humans, could produce drugs that can be repurposed for this pandemic much faster and be available for patient use.
REFERENCES
- Hameroff SR. Ultimate computing: biomolecular consciousness and nanotechnology. Elsevier; 2014.
- Miramontes P. A cellular automaton model for the evolution of nucleic acids. Thesis in mathematics; 1992.
- Danchin A, Medigue C, Gascuel O, Soldano H, Hénaut A. From data banks to data bases. Res Microbiol.1991;142(7-8):913-6.
- Sieburg HB. Physiological studies in silico. In 1990 Lectures in Complex Systems. 2018:367-390).
- Lipinski CA. Lead-and drug-like compounds: the rule-of-five revolution. Drug discovery today: Technologies. 2004;1(4):337-41.
- Oprea TI, Davis AM, Teague SJ, Leeson PD. Is there a difference between leads and drugs? A historical perspective. J Chem Inf Comput Sci 2001;41(5):1308-15.
- Pugazhendhi, D., Umamaheswari, T. (2013). In-silico methods in drug discovery-A review. Int J Adv Res Comput Sci Softw Eng. 3:680-683.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403-10.
- Durrant JD, McCammon JA. Molecular dynamics simulations and drug discovery. BMC Biol. 2011;9(1):1-9.
- March-Vila E, Pinzi L, Sturm N, Tinivella A, Engkvist O, Chen H, et al. On the integration of in silico drug design methods for drug repurposing. Front Pharmacol. 2017;8:298.
- Erlanson DA, Davis BJ, Jahnke W. Fragment-based drug discovery: advancing fragments in the absence of crystal structures. Cell Chem Biol.2019;26(1):9-15.
- Lin X, Li X, Lin X. A review on applications of computational methods in drug screening and design. Molecules. 2020;25(6):1375.
- Seidel T, Bryant SD, Ibis G, Poli G, Langer T. 3D pharmacophore modeling techniques in computer-aided molecular design using LigandScout. Tutorials Chemoinform. 2017;281:279-309.
- Sastry GM, Dixon SL, Sherman W. Rapid shape-based ligand alignment and virtual screening method based on atom/feature-pair similarities and volume overlap scoring. J Chem Inf Model. 2011;51(10):2455-66.
- Saxena S, Meher K, Rotella M, Vangala S, Chandran S, Malhotra N, et al. Identification of SGLT2 inhibitor Ertugliflozin as a treatment for COVID-19 using computational and experimental paradigm. bioRxiv. 2021;Jan 1.
- Saxena S, Meher K, Rotella M, Vangala S, Chandran S, Malhotra N, et al. In silico and in vitro demonstration of homoharrintonine antagonism of RBD-ACE2 binding and its anti-inflammatory and anti-thrombogenic properties in a 3d human vascular lung model. BioRxiv; 2021.
- Saxena S, Meher K, Rotella M, Vangala S, Chandran S, Malhotra N, et al. tyrosine kinase inhibitor family of drugs as prospective targeted therapy for COVID-19 based on in silico and 3d-human vascular lung model studies. BioRxiv; 2021.
Citation: Voleti S, Saxena S, Saxena U (2021) Emergence of New Drug Discovery Paradigm using Computational and Experimental Biology: Case Study of using Repurposing Drugs for COVID-19. J Infect Dis Diagn.6:152.
Copyright: © 2021 Voleti S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.