PMC/PubMed Indexed Articles
Indexed In
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- China National Knowledge Infrastructure (CNKI)
- Electronic Journals Library
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- MIAR
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Abstract
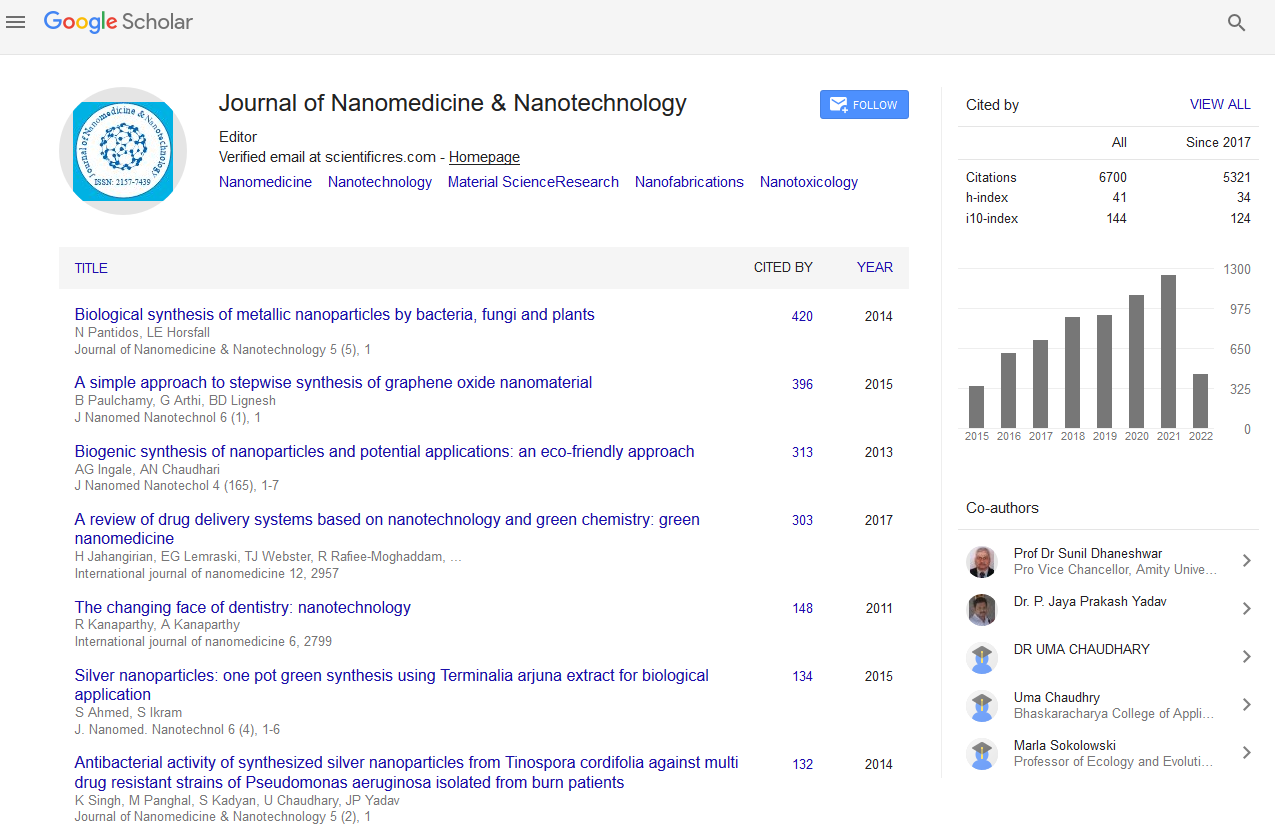
Diversity of Hydrocarbon-Related Catabolic Genes in Oil Samples from Potiguar Basin (Rn, Brazil)
Leandro Costa Lima Verde, Tiago Rodrigues e Silva, Bruna Martins Dellagnezze, Eugênio Vaz dos Santos Neto and Valéria Maia de Oliveira
Biodegradation may result in physicochemical changes in crude oil and natural gas properties, being responsible for the decrease of saturated hydrocarbons and yielding heavy oil with low economic value. Studies on the diversity of microbial catabolic genes in oil reservoirs are scarce and could help to predict the potential of a petroleum sample to be biodegraded. The aim of this study was to evaluate the diversity of genes involved in hydrocarbon degradation in Brazilian petroleum samples (biodegraded and non-biodegraded) through the construction and analysis of gene libraries (alkane monooxygenase – alk, dioxygenase – ARHDs and 6-oxocyclohex-1-ene-1-carbonyl-CoA hydrolase - bamA). The results showed a differential distribution of catabolic genes between the sites, being the biodegraded oil more diverse for the alk and bamA genes. Sequences were similar to the alkB genes from Geobacillus thermoleovorans and several species of Acinetobacter, to ARHD genes from Pseudomonas spp. and two species of Burkholderia, and to bamA genes from deltaproteobacteria. Interestingly, most of the catabolic sequences recovered from both petroleum reservoirs grouped together forming distinct clusters in the phylogenetic tree reconstruction and may correspond to potentially new genes, possibly harbored by yet uncultivated microorganisms. This is the first report on the detection of alk, ARHD and bamA genes in petroleum reservoir environments, demonstrating the genetic potential of such microbial communities to biodegrade the oil.